After extension, I need to take it out and put on ice box, and then open the lid to add EDTA. Then put back the membrane and seal it. Then stir and mix. Then spin down. This process takes another 30s or so.
In total, the error in terms of time measurement is ~1min, not including inaccuracy in temperature measurement. If you look at the time course curves, the data points at <1min are always very messy. That's because the time I'm measuring is smaller than error. If we further decrease the time interval, we will only get messier data.
Please also be advised that means time interval means more time points, more labor, longer time and higher reagent cost.
RC: Yes, I'm aware of the difficulty of higher rate sampling and I noticed the issues with those data points. However, you should not assume that 5 s sampling is the only improvement that can be made to the protocol (or that 5 s is the recommended value, since I may not have accurately recalled the precise number recommended). You should discuss to determine what the implications of the simulations are for the experiments, since the usability of the data relies on certain assumptions being satisfied, and it is better to know earlier what these implications and assumptions are. The simulations will be part of the paper, so a consistent story must be generated. It is also possible that the latest conclusions from the simulations has changed since I last received an update, and the issue with accurate initial rate estimation is less severe than I was initially told. I am conveying this information now so you do not find later that you did a lot of work that has to be repeated.
RC (2/19): Some of the recent simulations suggest that initial rate measurements may be advisable at 5 second intervals during the initial stage of the reaction (i.e. sampling more finely, if possible, at that stage, to get a better initial rate estimate). Since this may affect the experiments you are running/just ran, you can more information about this from Karthik who is available to connect again on the extension paper issues that you and he were discussing. Since both of you may have other work underway you can arrange a mutually convenient time to share info; just bear in mind it may affect your experiments.
Raj
CJ (2/10/14)
For Taq polymerase, NEB recommends 0.5–2.0 units per 50 µl reaction, ideally 1.25 units/50uL. The Specific Activity of Taq Polymerase = 292000 units/mg, which means 1 unit = 0.036 pmol. So 1.25U/50uL = 0.9nM.
RC (2/6): Please provide the typical enzyme concentration in PCR in nM units to Karthik.
CJ 1/27/2014
Attached please find a 'step-by-step procedure' for building a user-defined equation using Prism.
Some notes:
- I did not encounter any bug or problem. So no debugging information can be provided.
- There is no guarantee that given any equation and data, I can work out the curve fitting within 1-2h. The time needed varies from case to case.
- I’m following a built-in equation, so I know how to setup the initial values and constraints. With a new equation, we can start with the same setting. But I will need to fine tune the initial values and constraints before getting satisfactory fitting. This part of work MUST BE DONE WITH APPROPRIATE DATA. Without data, it is not possible for me to predict what initial values to use for each parameter in each different equation.
CJ 1/16/2014
About the processivity data: as mentioned in Raj's recent manuscript:
Literature data on Taq processivity at 72 C: E[ioff]=22 (reference: Wang et al., A novel strategy to engineering DNA polymerases for enhanced processivity and improved performance. Nucl. Acids Res. 32: 1197-1207).
The corresponding microscopic processivity for Taq is 0.95.
The experimental condition for this result is:10mM Tris-HCl pH8.8, 2.5mM MgCl. 50nM template, 0.005-0.1nM Taq, 250uM dNTP.
They did not vary [N] so it is hard to tell whether pocessivity depends on [N] or not.
Do we need more processivity data from literature? If so are we only interested in Taq, or also other polymerases? Knowing these information would save us time on literature searching.
RC (1/16): We're mostly interested in Taq, but we'd like to know whether there is any info regarding the effect of reaction conditions on processivity, since that would reveal something about the dissociation mechanism (as indicated in the notes). If you have already concluded that there is no information about this, or that processivity assays are always carried out under standardized conditions on an enzyme-specific basis, please let us know. In any case, the above info will be useful to Karthik in determining k-1 using processivity data.
Regarding Raj's question: RC: I believe you had a good linear fit for kcat/Kn(T)?
CJ (1/16/14): I never did linear fit. Before we toss in Sudha's old data, I did nonlinear MM fitting to get kcat and Kn. Those curves are well fitted. Then we added Sudha's data and did two variable nonlinear fit. For the reason I explained before, this fitting did not work well.
RC: I meant the Arrhenius fitting shown in the paper for the temperature dependence, with regarding to the question on whether we need to do new experiments varying [N] at 72C in addition to the ones already done at 70C. In the context of the two-variable fitting, you can consider this and let me know what you decide.
CJ(1/16/14): I see. So before we added Sudha's data the Arrehnius plot works well. After that, it is hard to say because I did not get any reliable parameters. And there's only 4 temperature points (instead of 6 as used before) that are common for Sudha's and my data. I'm not sure whether the Arrehnius plotting comming from single-variable fitting can be useful in two-variable fitting.
CJ 1/15/2014
I've read through Raj's posting on 1/13/14 including the attached file. I generally understand what we need to do in the next step. I will collect literature record on processivity and corresponding conditions and prepare a report hopefully by the end of this week. Meanwhile I have a couple of questions regarding the file Raj uploaded on 1/13/14.
(1) On page 1, it says 'Plot p_off(i) vs i; log p is slope. ...' I think it should be log(p_off(i)) that is to be plotted vs i.
RC: Yes.
(2) On page 4, second paragraph from top, it says 'We consider two CT systems that can produce the observed distribution..." What is a CT system?
RC: continuous time system - ie one framed in terms of reaction rates - as opposed to discrete system that was studied in previous treatments of processivity (e.g. by von Hippel). These systems allow us to get relative or absolute rate constants based on processivity data.
(3) On page 7, equation (1), I'm not quite sure what is (E.Di+1)'. People usually use ' symbol to indicate a closed conformation of polymerase-template-dNTP complex. After nucleotide incorporation, this complex will become polymerase-template-PPi. Then there will be a conformational change to come back to open state before ejecting PPi. After PPi leaves, the complex would remain in open conformation and therefore is usually indicated as (E.Di+1) without the prime symbol. Based on this mechanism, there should be no such species as (E.Di+1)'.
RC: I used this symbol before receiving the papers which used it to denote the closed conformation. I used it to refer to the intermediate state after nucleotide addition and prior to translocation. In these simplified models I did not include PPi dissociation. As noted the model with translocation may need to be modified based on the latest literature information, in particular the relative magnitudes of the various rate constants. The model I used was based on a reading of von Hippel's description of translocation.
(4) On page 7 at the bottom, there's a redundant paragraph. You may want to delete it.
(5) On page 15 at the bottom, you say Keq,1 should be replaced with an expression with k1, k-1, kcat, Kn etc. k-1 can be derived from k1 and Keq,1, I understand this. But how do we get k1? We use literature value? Or it is an additional parameters that needs to be determined by curve fitting?
RC: It is determined by curve fitting using time series data of the type you have obtained, as I believe may have been discussed in the parameter estimation section. This applies in the absence of processivity data. In the presence of processivity data, the MM fitting should give all unknown parameters.
(6) You ask me and Karthik to meet to discuss about the curve fitting using different models. As I have explained, our current data set does not work well with any model. In such case what should we discuss during the meeting?
RC: The strategy for going forward and subdivision of labor should be discussed. You should tell him about your literature review on mechanistic models. You should also tell him about the conditions under which processivity is used measured as noted below. Also, Karthik can use processivity data to predict the enzyme dissociation rate constant under model 1, and hence combine this info with Keq(T) from Datta to get the association rate constant as well. Based on these he can simulate the whole system. He can test the validity of MM steady state assumptions with this model as well.
(7) For additional experiments, do we want to do it at 72C (this is where we have processivity data) or 70C (we already have data with fixed template concnentration for this temperature, but not for 72C)?
RC: I believe you had a good linear fit for kcat/Kn(T)?
CJ (1/16/14): I never did linear fit. Before we toss in Sudha's old data, I did nonlinear MM fitting to get kcat and Kn. Those curves are well fitted. Then we added Sudha's data and did two variable nonlinear fit. For the reason I explained before, this fitting did not work well.
CJ 1/14/2014
In the attached spreadsheet is a list of DNA polymerase kinetic parameters I found on literature. The 5 references are also attached, 1 reference is missing because we do not have access to full text. The rate constant k1 - k8 are explained in the generic model pasted on the right side of the list. The models in the six references are mostly the same. None of them considered the translocation as a separate step. Experimentally, rate constant for translocation is combined with k6 and k-6.
kinetic parameter in literature.xls
Zahurancik 2013.pdf
I also found a couple of recent papers discussing the translocation as a separate step. All of them used single-molecule technology and their models are substantially different from the one discussed above so I did not include their results in the spreadsheet.
Lieberman 2013.pdf
Wang 2013.pdf
Maxwell 2013.pdf
The Wang 2013 paper may be of particular interest because they observed that during translocation, there are two states, one weakly bound and one strongly bound. This finding may be related to the salt-dependent and salt-independent states in Hippel's review.
RC (1-14): The bottom left para on pg 3881 on Wang is similar to some of the proposed approaches to parameter estimation made in the document I uploaded below, where we propose using equilibrium data (t=infty) to get processivity parameters and then use time series data (other t) like that you have obtained so far to estimate other parameters. Please pass this info to Karthik.
It will still be useful to know if there are some universal conditions under which processivity is measured, vis-a-vis the comments in the notes below on the different dependence of processivity on reaction conditions predicted by different models.
I will read Raj's latest post in details later today and tomorrow. Now just one thing I want to bring to our attention: using the current data by Sudha and me, the bireactant curve fitting would not work well no matter what model or software we use. The problem is that the two sets of data do not agree with each other quite well due to difference in experimental conditions. Below is the xls file I sent to Raj last week. If you open it and turn to sheet '65C Prism', look at the table in blue. If you go through row 53, you see the initial rate increases along with [SP], so in cell J53 we expect to have a number >146. If you go down through column J, you also see the initial rates increases with [N], so at cell J53 we expect to have a number between 62 and 72. This shows how the two group of results can not be integrated, although I have done all the possible adjustment to compensate for the difference in experimental condition.
bireactant model.xls
This is a common problem with data at all 4 temperature points. This problem can not be solved by changing the model or manipulating the data. If we really want to use the bireactant model to get good results, I'm afraid we will need to repeat Sudha's part with new protocol.
RC (1/14): Ok, this could be done at one temperature to start, after the discussion with Karthik on the priorities for next steps.
RC 1/13/14
polymerase kinetic models and parameter estimation results.docx
Attached is a polymerase modeling and parameter fitting summary that shows
how to get rate constants from processivity data, under different assumptions.
I have asked Karthik to consider simulating the simplest model (no translocation)
and comparing the results to your experimental time series data. In the meantime,
it will be helpful to get data on rate constants for the various steps in the full models
presented in the literature in order to determine the validity of the assumptions in the various models
we are considering in these notes. Also, we would like to get more information on the conditions
under which processivity is determined, because according to the different models considered,
processivity will depend on different parameters, as discussed in the notes. Hence, understanding
from the literature what factors processivity depends on will help us choose a model.
It may be necessary for you to work together with Karthik
to modify the models until you find the minimal model that fits your data. After you have provided the requested
information, I may ask you to start working together on this.
Here are some general comments:
1) The assumptions are used to justify various possible simplifications of
the full reaction scheme for polymerase extension, for example due to certain steps being
much faster than others or the steady state assumption for the nucleotide addition step being invoked.
Even so, none of the models presented account for all the steps in
extension. Recent work on polymerase mechanisms use around 6 reversible
steps in the full reaction scheme.
2) For PCR applications, we care most about the ability to predict the
extension time at any given temperature. Any aspect of the model that
does not affect this time significantly can be ignored.
Also, the following experimental methods and estimation techniques are
practical to apply in rapid thermostable polymerase characterization for
PCR modeling:
a) steady state kinetics (initial rates)
b) processivity
c) parameter estimation using time series data
Of these, we would only consider doing a,c) ourselves.
It is not very practical to do (pre-steady state and burst) kinetics
experiments that are done by labs that specialize in polymerases, when
characterizing various
thermostable polymerases at different temperatures for PCR applications.
3) The simplest model is the one you also considered without
translocation. There, we can use the method described in the notes to get
the dissociation rate constant
at 72C based on processivity data. KM will do this first. He can get
the binding rate constant as well using Datta and Licata's Keq(T). The MM
kinetics for this model shows that Keq(T) for polymerase binding is one Km
that can be obtained from fitting the MM data. For this model it will help to know the conditions under which processivity is measured. For example, does
processivity depend on [N]? The simplest model in the notes indicates that it does depend on [N].
4) If the model 3) does not predict the data well, it may be due to
various assumptions made. More complicated models can be considered. One
of these is the model with translocation. Here, as mentioned in the notes,
there is some ambiguity as to whether polymerase dissociation can occur
during translocation. The model I presented assumes that it can, and that
translocation occurs much faster than nucleotide addition. In this case,
processivity does not depend on [N]. Also, the dissociation rate constant
is not the same one that would follow from Datta and Licata's experiments,
since their experiments did not consider translocation. As shown the Km's
from the MM model have different interpretations in this model. This model
could also be tested against the data. Other, more complicated models can
be considered based on the same principles presented.
Note that the model I presented with translocation was based in part on von Hippel's commentary
on salt dependence of processivity. Based on the latest literature, this may not be correct.
But this model introduces some principles regarding how assumptions regarding certain steps
in the reaction mechanism being much faster than others can lead to model simplification.
This is related to the issue of the relative magnitudes of the rate constants in the full extension models
presented in the literature.
6) One concern I have with some of the simple models is the assumption that steady-state kinetics applies
throughout the reaction for nucleotide addition. This may be source of errors in the models. That assumption could be relaxed in the extension
paper, if the estimation methods suggested are used.
*7) If the extension paper is to focus on building and testing predictive models for extension time
calculations, it will be necessary to consider more than one model and
numerically implement several estimation schemes. These estimation schemes
would involve MM model fitting, as well as the use of time series data for parameter estimation, as discussed in the notes. The models would need
to be simulated and the predictions compared to experimental time series
data until a suitably accurate minimal model is found. It would be
necessary for two team members to work together on these issues. Some of the underlying principles have been provided in the notes.
I may post a full version of the notes above including more details on how the results were obtained shortly. These details are not required for the proposed
work at this time.
Raj
CJ 1/10/14
Regarding your comments on 1/6/14:
1. I'm not sure what did you refer to by saying 'some of the literature (eg., Benkovic, Johnson)'. Would you please attached the two papers you mentioned? I found Kenneth A. Johnson's 1993 review on DNA polymerase conformational coupling. However, I did not see him mentioning 'dissociation is assumed to occur prior to nucleotide addition'.
RC: I was referring to the assumption that enzyme dissociation occurs as the reverse reaction of enzyme binding to the primer template complex in the first step of the polymerase reaction scheme, but is not considered to occur during translocation.
2. There are some serious mistakes in Peter H. Von Hippel's review 'on the processivity of polymerases'. In equation (5) he said the nT is defined before. But actually it should not be the nT defined before. Instead nT in equation (5) should be total density of all bands. The paper by Yan Wang on NAR 2014 cited Hippel's review but corrected this mistake. Also on Hippel's review, page 129, the last paragraph in main text, he said that 'Polymerases of different types may react differently to template "obstructions" that are characterized by high PI values'. This statement seems to be wrong. The obstructions on template should be characterized by low PI values. With these critical problems I would be very cautious with any conclusion in this paper.
3. The Hippel's review also cited a lot of unpublished results (especially for the salt-dependency studies) from his own group, making it very difficult to verify those conclusion. Considering both 2 and 3, I'm not sure how reliable this review is.
4. Below I'm posting three original paper published in 2013. There are a lot of things going on since Hippel's 1994 review. Hopefully these new papers would bring some inspirations.
ja403640b.pdf
bi400803v.pdf
ja311603r.pdf
Regarding your comments today:
1. I will look into the different rate constant of different DNA polymerases. One thing to clarify: are we only interested in those polymerases commonly used in PCR, right? There are also a lot of studies on DNA polymerases in human or viruses that are not used for PCR, do we also want to include them?
RC: If data for PCR polymerase is available it's fine, but since the only data that was presented to me in the past regarding enzyme binding and dissociation rates were for non-PCR polymerases, I didn't know whether PCR polymerases were as exhaustively characterized kinetically.
2. In Prism it is doable to input user-defined equation. I tried it a little bit but found it takes some time to debug. But if we really need to deal with it, I can try more.
RC: It will be important to determine if this is possible and if so how difficult, in the near future, so we can decide whether to use this software or not.
CJ: We can use this software.
CJ (12/27/2013)
Attached below is the reference you are looking for. I will also read it.
j.1749-6632.1994.tb52803.x.pdf
About the processivity assay, it is not hard, but it really relies on DNA sequencer, or CE with fluorescence detector. Basically, they label primers with fluorescent dyes, and run extension. Then the extended products are loaded to DNA sequencer. DNA sequencer for Sanger sequencing basically runs capillary electrophoresis and then detects products based on their fluorescence. In this way they can know the concentration distribution of extension product at different lengths (the concentration is measured by fluorescence). Compared to PAGE gel, CE is more accurate for quantification and more sensitive (which means low detection limit and also it can distinguish two strands with one single nucleotide difference).
RC (1-6): I would like you to look into an issue regarding the mechanism of polymerase dissociation from the primer-template complex. There appears to be some ambiguities in the literature regarding when the polymerase dissociates. In some of the literature (e.g., Benkovic, Johnson) including some references cited in the above paper, dissociation is assumed to occur prior to nucleotide addition. In the above review, it is stated that polymerase dissociation that is responsible for processivity is highly dependent on salt concentration, since electrostatic interactions are primarily responsible for maintaining template binding during the translocation step of polymerization that occurs after nucleotide addition, when the polymerase moves to the next position on the template. This seems to imply that most of the dissociation occurs during the translocation step. However, dissociation during translocation does not appear to be addressed at all in the earlier literature. Please investigate whether any other literature has mentioned dissociation during the translocation step. Also, please check the standard conditions (concentrations) under which processivity is measured. For example, are measurements always made under saturating nucleotide concentrations? And at a specific salt concentration? Please also make a list of all known rate constants (including translocation rates) and processivities of a few well-studied polymerases and post to the wiki. At this time, please investigate these questions only as we have various other modeling efforts underway that address other issues. I will provide a detailed document summarizing the modeling efforts shortly.
t7 polymerase mechanism.pdf
RC (1-10): I have prepared a summary of the modified MM theory that accommodates various possible kinetic schemes - including translocation/dissociation during translocation and omitting a treatment of translocation. There is also a discussion of how available processivity data can be used in our fitting. I will be posting that by Mon. The rate constants for the various steps for a couple of polymerases will be helpful in choosing among the kinetic schemes. Hence in this document I will not be specifying one particular scheme to use, but rather presenting various options.
It will also be helpful to know whether entering an arbitrary MM multivariable model equation into Prism is very difficult.
RC(12/16): Please read the methods section of the attached paper and comment on how difficult it would be to do this processivity assay in our lab. Also, please look up and post reference 27 (von Hippel).
processivity.pdf
CJ 12/19/13
Here I'm attaching 7 xls files. The first one is a summary of fitted dsDNA concentration at all temperatures, all [N]0, and all time points. Note that [E]0 and [N]0 vary from one temperature to another, as indicated in each sheet.
121913 fitted curve summary.xls
Below are 6 xls files of the fitted concentration vs. experimental data, as well as the information of the curve fitting, and fitted data at more time points, each file for one temperature.
The file for 75C is slightly different with what I posted last time. I corrected some errors during copy and paste.
121913 55C fitted curve.xls
121913 50C fitted curve.xls
121813 70C fitted curve.xls
121813 65C fitted curve.xls
121813 60C fitted curve.xls
121713 75C fitted curve.xls
CJ 12/18/13
Attached below are two files with XY values of fitted time course for all dNTP concentrations at 75C. The pdf file is a summary of fitted values compared to experimental values at 12 experimental time points. The spread sheet includes full results at 1000 time points between 0-10min. The xls file also includes curve fitting parameters and raw data in terms of RFU versus minutes. In the pdf file the units are converted to nM versus seconds using calibration curve.
121713 75C fitted curve.pdf
121713 75C fitted curve.xls
I will work on other temperatures in the following two days.
CJ (12/17)
Sorry for the missing attachment. I'm attaching the spread sheet here.
Keq for Raj.xls
CJ (12/16):
Just want to clarify on three things:
(1) By saying 'Concentration values from fitted curve ...', you are referring to concentration of what? Total dsDNA? Anything else?
RC: ds nucleotide since that is what is being measured.
(2) You want this concentration values for all temperatures, all dNTP concentrations, and all time points? That means 6*12*12 = 864 numbers in 72 curves. If so, please allow me some time (2-3 days) to finish them.
RC: All times for the highest few temperatures (e.g. 65,70,75) for 1000uM would be needed. This can wait for 1-2 days.
(3) By saying 'Keq for enzyme binding', are you referring to dissociation constant of enzyme with template (Keq,1)? or enzyme with dNTP (Kn)? The former is taken from Datta and LiCata paper and I have sent them to you on Friday by email. The latter is reported in the manuscript.
In case you did not get my email on Friday, I'm attaching the Datta and LiCata paper here:
Thermodynamics of the binding of Thermus.pdf
RC: Keq,1. The email from Fri said "Attached is a spread sheet with Datta and Licata's Kep data. " but there was no attachment.
You mentioned you extrapolated some values.
Currently I'm working on the sequencing results of the beta-lactamase library. I will come up with a report by tomorrow. After that is done, and after all the points here are clarified, I will come back to the paper.
RC (12/16):
Chaoran,
Please provide the following, the first three of which I need for some calculations I am doing:
- Concentration values from fitted curve at each time point for all curves in your ppts. You can repost all your ppts on the wiki with these values indicated.
- Any info on the fitting provided by prism.
- Keq for enzyme binding at each temperature
CJ 12/11/2013
Regarding Raj's comemnts:
RC:Is the time series data with fitting at a particular temperature available in the manuscript for 1000uM? If not please post it here. Please also tell me the concentration of nucleotide at the last time point at a particular temperature (say 72 C).
CJ: Please see a report in the attached slides.
121113 for Raj.ppt
(I updated the attachment.)
RC: I mean whether the aforementioned rate measurement can be made in real time during a real-time PCR reaction. I would assume this is difficult. As you can see from the equations presented, it is not just a matter of total dNTP incorporation after each cycle.
CJ: I agree with you.
RC: If we don't include the simulation we will need to move some of the MM derivations into the body. This will be the next step after the issues above are settled. After these changes the paper should be put in journal format (probably NAR), even if we choose to later put in simulation content. What are your thoughts on content vis-a-vis NAR standards if the simulation content is removed. I will also consider this.
CJ: I see. For NAR, I feel that we'd better add some simulation work to match its high profile (IF 8.28).
CJ 12/03/2013
Attached below are four files:
(1) a near-final manuscript with track change.
Taq Paper CJ 11272013.doc
(2) a draft of Supporting Information. I temporarily moved the simulation work in SI.
Taq Paper SI CJ 120213.doc
(3) my updated comments.
CJ comments 11272013.doc
(4) a sample paper on NAR.
Thermodynamics of the binding of Thermus.pdf
RC: I am copying the remaining two questions here:
8) Can accurate fluorescence measurements be made under pseudo first order conditions (higher [N] excess)? If these conditions cannot be not used, we must use numerical simulations to make the predictions.
RC: The ratio of [N] to [SP]_0 is relevant. In early stages of PCR, the template concentration is lower, I think, than that used in our experiments. With higher template concentration (lower ratio), nucleotide depletes more quickly. I am curious about the scope for changing this ratio from an experimental signal-to-noise standpoint. The ratio is low enough in the current experiment that the reaction cannot be considered pseudo-first order in the E.Di, since [N] clearly changes (see below) during the time over which measurements are made. The measurements in the current experiments also appear to be made sufficiently early such that the \sum_i=0^(n-1) [E.Di] remains roughly constant, as evidenced by the first order kinetics in [N], whereas we are interested in measurements at later times when a lot of full-length DNA is being formed.
CJ: I’m still not quite clear on what you want me to provide here. Again, I don’t see any reason why high [N] would make the measurement inaccurate. I have never observed a deterioration in accuracy at high [N] (up to 1000uM) in experiments, either.
RC: So you believe that [N]_0 can be raised sufficiently high that [N](t) is approximately = [N]_0 even when most of the [SP]_0 has been converted to DNA, without compromising the measurements. If so, you should provide the maximum [N]_0 that you feel would be viable, and then indicate the [N] when all [SP]_0 has been converted to DNA. Based on this I can assess the accuracy of the pseudo-first order approximation and hence the suitability of the simulation method to the experimental setup.
CJ 12/11/13: the highest [N]0 in our experiment is 1000uM. The [N] consumed in the extension is 0.2uM X 63 = 12.6uM (0.2uM is [SP]0; 63 is the length of ssDNA in SP complex). So by the end of the reaction only 1.3% of the [N] is consumed. Under such condition I did not observe any deterioration in data quality.
RC: Ok, this could be a suitable pseudo-first order condition, though we may increase [N] further beyond this to improve the approximation. I assume a 5-10x increase in [N]_0 would be ok. Is the time series data with fitting at a particular temperature available in the manuscript for 1000uM? If not please post it here. Please also tell me the concentration of nucleotide at the last time point at a particular temperature (say 72 C).
9) A method for determining concentration of fully extended DNA at any time based on solution phase fluorescence measurements is proposed. It based on a rate measurement. Are these measurements inaccurate without a sufficiently large number of measurements to obtain the slope. Would the required reaction conditions decrease signal to noise due to background fluorescence? Would the signal to noise during PCR cycles be too low to use this method to obtain the final DNA product concentration? If not, we may portray this as another application of our experimental work, since it could be used in PCR without model simulations
RC: The method described on pg 26 under comment RC23 (where this comment is taken from). It is based on rate measurement under conditions of sufficiently high nucleotide excess that the d/dt[N] approximately = 0, and where the sum of intermediate concentrations is not constant - unlike the current experimental protocol, which displays first order kinetics with respect to [N]. The protocol would otherwise be analogous to the rate measurements in the current experiments.
CJ: I am still not quite clear on exactly what is the ‘method for determining concentration of fully extended DNA at any time’. Do you mean the gel-based assay? Without running a real experiment, it is hard for me to predict whether the S/N of a gel-based assay would be good enough for monitor the procession of PCR or not.
RC: As described using equations in the manuscript, the method is to measure the rate of nucleotide incorporation. Since this rate is proportional to the sum of all [E.Di] concentrations, i<n, it allows us to determine \sum_i=0^n-1 [E.Di] at any time and hence by mass balance calculate DNA concentration [E.Dn], assuming we know kcat/Kn. This is a solution measurement of rate like the ones done for this paper, but we may want to do them under the pseudo-first order conditions mentioned above. Hence we need to know whether accurate rate measurements can be made under the conditions determined under 8).
How long would it take to do these experiments? I am also asking you to comment on whether these measurements can be made under standard PCR reaction conditions.
CJ 12/11/13: Our current protocol is quite labor-intensive. It takes two days to finish one set of experiments, which means measuring dNTP incorporation at 12 different time points, for a fixed extension temperature. Various reactant concentrations ([SP]0, [E]0, [N]0 etc.) can be assayed in one set of experiment. But there's a limit in the number of samples I can handle simultaneously. Currently I'm running 8 different [N]0 points in one batch, which is kind of the maximum. Would you specify how many time points, concentration points, and temperature points you would like to obtain, so that I can estimate the time needed.
RC: We would need only one [N]_0 and one temperature. The number of time points could in principle be smaller, since we do not need to fit a curve, but since we will be running fewer reactions we may in fact increase the number of time points. We may make some measurements at later times. Assuming for the time being that the number of time points is the same, how long would the experiment take?
The data could be used in two ways: a) comparison of absolute [N] at a particular time to the model prediction; b) comparison of the [dsDNA] obtained from the rate measurement method above (not an initial rate measurement, but rate at the specified time) to the model prediction. In the latter case it will be necessary to obtain an accurate rate estimate at -any- specified time during the reaction.
I'm not quite sure about the term 'standard PCR reaction conditions' you used above. If you mean the enzyme, template, dNTP concentration etc, our current assay is quite comparable to standard PCR reaction conditions. Or do you mean you want to monitor the dNTP incorporation during a regular PCR program? If it is the later case, why don't we use the qPCR method, which is designed to measure dsDNA concentration in each cycle of PCR program.
RC: I mean whether the aforementioned rate measurement can be made in real time during a real-time PCR reaction. I would assume this is difficult. As you can see from the equations presented, it is not just a matter of total dNTP incorporation after each cycle.
What else remains to complete this version? I will let you know whether we will be retaining some of the simulation parts, after assessing the answers to the questions above.
If we do not include the simulations, we will probably move some of the SI MM derivations back into the paper, possibly in an Appendix, since no one generally reads Supporting Info.
CJ 12/11/13: It is mostly done. We just need to fix the simulation part. If we move the simulation back, we will need to adjust the abstract, background, discussion and conclusion part accordingly. If we decide not to include the simulation part, all I need to do is to proof read the manuscript and fit it into the NAS template for submitting.
RC: If we don't include the simulation we will need to move some of the MM derivations into the body. This will be the next step after the issues above are settled. After these changes the paper should be put in journal format (probably NAR), even if we choose to later put in simulation content. What are your thoughts on content vis-a-vis NAR standards if the simulation content is removed. I will also consider this.
CJ 11/25/2013
Attache here is the updated version of the Taq extension manuscript. I am also attaching a separate file to answer Raj's questions. I need to spend this afternoon and tomorrow morning to finalize the slides and prepare for the talk on group meeting. I will finish the rest of the revision after the group meeting.
Taq Paper CJ 11212013.doc
CJ comments 11252013.doc
RC: Some replies attached.
CJ comments 11252013_RC.doc
CJ 11/20/2013
Raj,
I read through you new MM model and have a question on Keq. Please see my comment on the attached file.
question.pdf
RC: Since the approach to calculation of Keq based on our own data would require us to use Sudha's data, I was not planning to apply that equation here (given the issues you raised with Sudha's data), but rather simply provide it to show that we have developed a methodology to do so. The contribution of our paper is a method for any polymerase as well as our data for Taq polymerase. (We made some arguments as to why we expect the effect of uncertainty in Keq provided by Datta - since it is not for Taq - to have a small effect on our calculated Kn, based on our chosen reaction conditions.)
RC (11-14):
Chaoran,
Attached below are my latest revisions to the extension paper draft.
I am summarizing here the next steps that you can work on. These are also listed as comments in the working draft:
1) Length: Some parts of introduction are too long. Compare current length to admissible journal paper length and reduce as appropriate. Note that I have added the MM derivations to the body of the paper, since it is useful to have a unified model on which our experiments as well as new experiments can be based.
The appendix contains supporting derivations that are not essential (it is not necessary to read all these at this time). They could be moved to supporting information/eliminated, or some can be summarized and incorporated into the body of the paper after we finish all other tasks.
2) We should redo the MM calculations using equation (6) for 1/v. The differences with respect to the original single reactants formulation originate in the expression for [E] in terms of [E]0 . The original formulation considered partitioning into two intermediates separately (not exact, since formation of the 2nd intermediate shifts the 1st equilibrium, to an extent that depends on the steady state concentration of the 2nd intermediate; here we of course assume the 2nd intermediate cannot dissociate directly to E + S1 + S2). The expression for 1/v changes as a result compared to the original single reactant formulation. The changes are:
a) A correction term 1/ (Keq,1*[SP]0) that was previously neglected
b) The value of [E]0 used - previously, I believe [E]0 (which was the [E.SP]0 in the bireactants formulation) was calculated using [E]0 = [E.SP]0=[E][SP]Keq.
The approximations previously made may have been valid esp due to high SP concentration– we will see. In any case, the current formulation is preferred since it does not make as many assumptions. Note that the standard sequential bireactants derivation makes a rapid equilibrium assumption that we cannot make in our work, since we do not want to equate Kn with an equilibrium constant.
Related, the current protocol does not clearly explain how the enzyme concentration was chosen – where is Datta and Licata cited?
3) I have made a comment regarding why one must be careful in setting up MM experiments to determine Keq for enzyme binding. You can decide whether to include this statement after considering it.
.
4) Related to 3), enzyme dissociation during extension may be related to polymerase processivity. CJ please look into processivity and comment.
5) In the results section, should there be any commentary on comparison to the fitting in the BP draft obtained from Innis et al data?
6) Overall editing of all sections except for simulation and robustness. Assume these sections and associated commentary in the discussion, conclusion, etc will not be included in the paper . Please aim to finalize the paper including all formatting so it can be submitted without those sections if needed. This includes finalization of conclusion.
7) Journal choice: Assume simulation content will not be included. Please check length and content and recommend a journal, including analysis of related papers in NAR.
The following comments/questions pertain to the experimental comparisons to the simulations that could be made. They are also provided as comments in the draft. Based on answers to these questions, I will decide whether it is worth finishing the unfinished simulation sections or leave them for another paper.
8) Can accurate fluorescence measurements be made under pseudo first order conditions (higher [N] excess)? If these conditions cannot be not used, we must use numerical simulations to make the predictions.
9) A method for determining concentration of fully extended DNA at any time based on solution phase fluorescence measurements is proposed. It based on a rate measurement. Are these measurements inaccurate without a sufficiently large number of measurements to obtain the slope. Would the required reaction conditions decrease signal to noise due to background fluorescence? Would the signal to noise during PCR cycles be too low to use this method to obtain the final DNA product concentration? If not, we may portray this as another application of our experimental work, since it could be used in PCR without model simulations
10) Please comment on other methods for the experimental measurement of fully extended DNA, both offline (e.g. gels) and online (e.g. probes).
11) Please use the expression [E.S1.S2]=Keq,1/Kn [E][S1][S2] to compute the total concentration of the nucleotide intermediate, according to the eqns provided. This is an application of Kn (not just kcat/Kn) and will help us determine whether omission of the intermediate can be justified in the modeling.
Thanks
Raj
Taq Paper CJ RC 11-14-13.doc
CJ 11/5/2013
Please see two sample papers attached, from Biochemistry (IF 3.4) and Analytical Biochemistry (IF 2.6), respectively.
analytical biochemistry sample.pdf
biochemistry sample.pdf
CJ 10/31/2013
Please see the attached file for the current version of the Taq extension paper, in track change mode. Compared to the last version I added two paragraphs in the Method section, and one paragraph and a figure on the temperature dependence analysis.
Taq Paper CJ 103113.doc
RC: Please provide some commentary below regarding suggested journals for this work in the event that we do not include substantial theoretical modeling (i.e., if the content is largely based on the current experimental analysis).
CJ: I suggest Analytical Biochemistry or Biochemistry.
RC: Please send the closest analogous paper you can find published in Biochemistry as an example along with impact factors for both the above journals.
CJ 10/30/2013
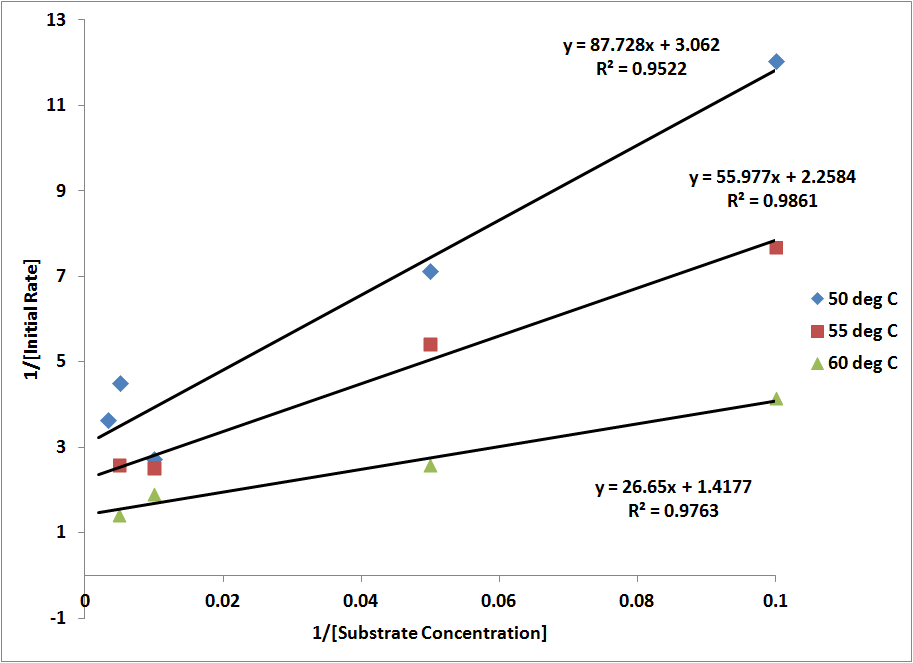
I checked Karthik's BP manuscript and found he plotted ln(kcat/Km) vs 1/T, instead of ln(kcat) vs 1/T. I tried the same thing and got the figure attached below:
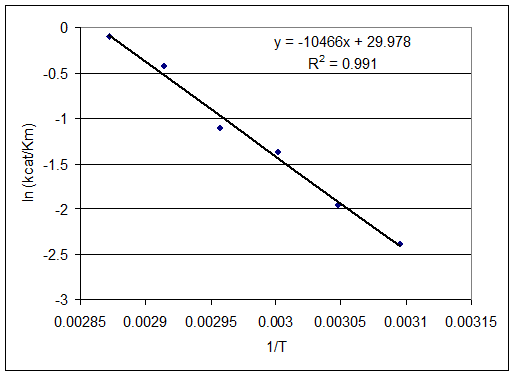
ln(kcat/Km) vs 1/T shows a linear relationship. However, this is no longer an Arrhenius plotting, and therefore the slope, as far as I understand, can not be used to derive Ea. I'm having a hard time finding the biophysical significance to this plot. Any suggestion?
RC (10-30): Yes, that's what I mentioned in my last posting this morning (see below). As noted below kcat/Kn is a second order rate constant that treats the reaction as E + S -> E + P, i.e., neglecting the ES intermediate in the model. You are right that it may not be appropriate to equate Ea obtained from this plot with an activation energy associated with a particular transition state barrier. However, the Arrhenius model (with its assumptions of temperature independent parameters) is typically only an approximation to transition state theory anyway. Apparently some of the temperature-dependent deviation of kcat from the Arrhenius model is canceled by temperature dependent changes in Kn. We do not need to explicitly indicate a relation between the slope and activation energy. Nonetheless the fact that we can use a single Ea/k0 is good for modeling purposes. I will comment more after we have incorporated this into the draft.
CJ 10/29/2013
See my reply below.
a) What was the fate of Sudha's work on Km determination for the DNA? Did you eliminate this because of the protocol she used? If so, what were the issues? Was the reason that a lot of the original manuscript was deleted?
In a sequential binding model (which means the Taq binds to template first, followed by dNTP), the Km measured in Sudha's way does not solely reflect binding affinity with the first substrate. It is usually reported as Km(app). Please see the figure below for a derivation on how Km(app) is different from real Km.
CJ (10-30) The figures were messed up. So I'm attaching the Word doc for a discussion on Km.
Km in sequential binding model.doc
The reason I deleted her figure is: (1) it's not easy to explain the biological relevance of Km(app). (2) Km for Taq and template has been reported using 'direct' assay like fluorescence anisotropy (Datta and Licatta). It is hard to explain why our method is better than theirs. and (3) the general protocol Sudha used for that set of data has been modified later, in terms of enzyme concentration, Mg concentration, time points and temperature points etc. Then we need to either explain why we use different protocols for template and dNTPs, or redo the whole set of experiments using the same protocol as for dNTPs. Either one would not be an easy task.
However, her Km values might still be worth reporting. I'm considering making it into the Supplementary Information.
RC (10-29): Yes, please consider it and update. In the meantime I am looking over your analysis above.
CJ (10-30): Please see the file attached.
Taq Paper SI CJ 103013.doc
RC (10-3): I think my comments from today were deleted. Yes, we considered this when we changed the MM protocol. Comments on this and how it should be presented will be forthcoming.
b) Where (what page) did you include the discussion about nucleotide inhibition?
After adjusting dNTP concentration, I did not see dNTP inhibition anymore. Therefor I find no need to discuss it. A discussion on dNTP concentration is in the last paragraph in Discussion section.
RC (10-29): If I understand correctly, you adjusted dNTP concentration because of inhibition and based on the known mechanism of inhibition through Mg chelation. Do you feel that emphasizing this would call into question the results/protocol?
CJ (10-30): Sorry for the confusion. I should say 'after adjusting Mg concentration'.
c) The robustness analysis (if included) will be done using the method different from (and perhaps shorter than) that you have included in the draft. I will revise that section. At the same time, I will decide on the content of the simulation section. How much of this we choose to include will affect the choice of journal.
I see.
d) What are some example journals you referring to when you indicate a 7000 word limit?
I used to publish on JACS, ACS chemical biology and Acc. Chem. Res.. They all have word limit of 6000 - 6500 for articles. I can check for the specific requirement once we decide on what journal it would go to
RC (10-29): Can you compare to the length requirements of journals like Nucleic Acids Research, Biophysical Journal, and PLOS Computational Biology (XG has info on some of these)? The latter are unlikely choices, but may be appropriate if we choose to include more simulations. Some of those may allow for longer papers. Do you feel any of the journals you mentioned are appropriate for this work?
CJ (10-30): Nucleic Acid Research charges excessive page fee ($195 per page) for papers over 9 pages (corresponding to ~7500 words). Biophysical Journal has a page limit of 10 pages, corresponding to 8000~8500 words. I have not found page limit information for PLOS Computational Biology yet. If we decide to submit to this journal I can email their editorial board to ask.
e) You did not appear to consider an Arrhenius model for extension rates as a function of temperature (i.e., a figure that examines whether a constant preexponential factor and activation energy can accurately predict the temperature variation of extension rates). Is this something you plan to add?
I used to ask this question in the previous round of revision, but got no answer.. Therefore I thought we were not interested in it. However I can try to add it. One thing we need to keep in mind is that enzymatic reaction may not fit well in Arrhenius model. In classic Arrhenius model, reaction rate simply increase with temperature. But for enzymes, reaction rate will peak at optimal temperature. If we do report the
Arrhenius thing, we will need to explain why it is significant.
RC (10-29): Even if we can apply a different Arrhenius model to say, two different temperature ranges, it is ok and useful for simulation. You can try to fit the model separately over 2-3 temperature ranges. In some cases we have found a good fit with multiple Arrhenius models when the reaction rate peaks at a particular temperature.
Significance/motivation: For computational modeling and optimization of PCR, it is necessary to have a model for the temperature dependence of all reaction rate constants. Knowing the rate constants at a discrete set of temperatures (such as those considered in the lab) is not sufficient since the optimization algorithm may vary the temperature and reaction time continuously. For example, since extension occurs even during the annealing step of PCR, we may need to know the extension rate constant at all possible annealing temperatures so that the optimal annealing temperature and time can be set. Arrhenius models are approximations, but they are convenient because of their use of just two parameters (preexponential factor, activation energy). We could fit various other nonlinear functions, but we should first examine the results with the simplest Arrhenius model. The goal is to show that such models for temperature variation of the rate constants are suitably accurate for the purpose of modeling and optimization.
You can talk to Ping about this, just for background on what we did previously with fitting of models for the temperature variation of rate constants. He is familiar with the work I mentioned and has the drafts (Biophysical J paper). This would help you outline this section. In return, I also asked Ping to talk to you about Datta and LiCata (enzyme binding rate constants). In the BP paper draft we may have a figure for variation of the second order extension rate constant kcat/Kn with temperature (an approximate model based on several assumptions and literature data, since we did not have the necessary experimental data at that time).
CJ (10-30): Please see the attached figure. Based on Arrhenius equation, if we plot lnk vs 1/T, the slope of the curve is -Ea/R. As you can see in the figure, lnk vs 1/T does not show a linear relationship. I'm not quite comfortable to use the multiple Arrhenius model because we only have 6 temperature points, barely enough to fit one model. If we want to estimate reaction rate at other temperatures, I suppose the best way is to fit all 6 data points into a nonlinear model, rather than 2 or more linear models. Another way is to do interpolation and extrapolation rather than curve fitting. What would you suggest?

RC (10-30): Such a figure should certainly be included in the paper. I should have been more specific; what we need for the purpose of modeling is the analogous plot for the second order rate constant kcat/Kn (ratio), since we do not model the intermediate. As mentioned above, we can fit other nonlinear functions (or use interpolation schemes) now that we have seen the data do not fit well to a linear model, but please discuss with PL first and get the plot we currently have in the BP paper (if it's there) and then we will decide on the approach (which approach chosen has implications for the modeling). In some cases we used two linear models if there was a good fit to the linear models with constant Ea,k0 in separate temperature ranges (e.g. when there was a temperature of maximal activity; here, we see maximal activity at the highest temperature you used, so that is not relevant). I agree regarding the current number of data points being limited in this case (have you sampled all temperatures you intend to)? (If we want more, we should discuss - it may be possible to get the data we need regarding kcat/Kn more rapidly.)
-- Robustness experiments. I've read your comments below. Here are some further details on how the experiments used for model validation (as opposed to MM parameter fitting) may differ. This list is in order of priority. Bear in mind that we may not do include the results of all such experiments for this paper (we may put them in a second paper instead).
a) We need to quantify the standard error of measurements of total DNA concentration for a known total concentration of DNA. This is because the difference between the model predicted total DNA concentration, given an estimated value of kcat/Kn, and the measured total DNA concentration is attributable to both the error in the model prediction (due to e.g. error in the kcat/Kn estimate) and the measurement error, and we are interested in the former.
I see. I will try to do some error analysis based on our current data set.
RC (10-29): Ideally this would be done with a known concentration of total incorporated nucleotide or DNA.
CJ (10-30): I'm a little bit confused on your terminology: you said 'We need to quantify the standard error of measurements of total DNA concentration for a known total concentration of DNA'. What is the difference between 'total DNA concentration' and 'total concentration of DNA'? My understand to 'concentration of total incorporated nucleotide or DNA' is that this is what we measured on the fluorometer, am I right?
RC (10-30): Same thing - just making sure the concentration is known so we isolate measurement error. To be precise, here and elsewhere I am referring to the fluorescence measurement of total ds nucleotide concentration.
b) Regarding the protocol suggested below, it is good to see that the conditions are similar to those in PCR. Please indicate how your lowest [dNTP] compare to those used in the later cycles of PCR (where nucleotide gets depleted). What are some characteristic values for the latter? The model can be used to predict incorporated [dNTP] under non pseudo-first order conditions as well, but we should first verify whether those are relevant.
Our lowest [dNTP] is 2uM. At this low concentration we can barely detect positive reactions (the value of initial rate is usually smaller than standard error). In late stage of PCR when dNTP is depleted, the dNTP concentration might be even lower, and the reaction rate is close to zero. As a result, I don't think it is very useful to study the kinetics under dNTP depletion. People are more interested in the initial and log phase of the reaction.
RC (10-29): It would still be useful to provide info on the characteristic [dNTP] at different cycles/stage of PCR since we ultimately will be modeling the PCR reaction (and providing prescriptions for the optimal temperature protocol) during every cycle (at least in other work). (It is interesting that you observe the reaction rate fall close to zero when [dNTP] is not in significant excess, and that you do not observe a regime where the reaction is second order (dNTP, template).)
CJ (10-30): If we want to know the dNTP concentration during each cycle, what we need to to is to run a series of PCR reactions, stopped at various cycles; measure the amount of incorporated dNTP and then calculate how much dNTP is left in the buffer.
Did you find that the first-order kinetic model fit the data equally well at your lowest vs highest [dNTP], indicating that a pseudo-first order approximation was appropriate at all [dNTP] concentrations?
At low [dNTP] and/or low temperature the first-order kinetics degenerates into zero-order: RFU vs. time shows a linear relationship. This makes sense because under these unfavorable conditions the reaction rate is so low that we can not sample all the way to the plateau stage within 10min.
While we may not change the protocol given that it suits PCR, we may want take more measurements of total DNA concentration at particular times to get a better estimate of the experimental uncertainty when comparing to model predictions.
I see. One thing is that the current protocols is quite time- and reagent-consuming, as well as labor-intensive, making it unrealistic to sample plenty of time points. I'm thinking of using a real-time PCR protocol to do this. That means we add the dye into the reaction and monitor the fluorescence over time. As discussed in the paper, this will introduce some complexity, eg, the dye may inhibit the enzyme. However, this is still worth studying because this is exactly what happens in qPCR application.
This is just some initial idea. If we insists sticking with the current end-point detection protocol, I'm ok with it. It just means we will need to spend a lot of time and reagents to get these data.
RC (10-29): We wouldn't need to repeat most of the previous experiments with more measurements at each time point. This could be done for only a few time points for one or two reaction conditions, since we would be making computational predictions at particular times.
CJ (10-30): My only concern is the day-to-day variation. Sudha used to observed that the exactly same reaction, run in 2012 and 2013, showed very different RFU values. This variation is not a big problem in our previous set of experiments because we ultimately use dRFU/dt rather than the absolute values of RFU. But for the experiments you suggested above, I think we need to make sure the results from our old data set is compatible with those from a new set of expt.
c) Though the current measurements can give us the total concentration of incorporated nucleotides, in order to determine the total concentration of fully extended DNA at a particular time (which the theoretical model can predict and which is of greatest interest in PCR), we may need to run a gel and extract the fully extended product. Please provide comments on how difficult this would be and the associated measurement error of the fully extended DNA concentration (esp how it compares to the fluorescence measurement error of the total incorporated dNTP concentration).
It is doable. But since detection of DNA on gel is also based on fluorescence, I don't see any reason why gel-based assay could be significantly more accurate compared to solution-based assay. The detector for solution-based assay is a fluorescence spectrometer; while for the gel-based assay it is a camera. My intuition is that the former would be more sensitive and accurate. The major advantage of a gel-based assay, in my opinion, would be the resolution of product of different length, if we are interested in. By the way, gel-based detection assay will be more expensive, time-consuming, and label intensive compared to solution-based assay.
RC (10-29): Yes I agree - I meant that I assume the accuracy of the gel-based measurement will be lower than that of solution phase measurement, and that is why I am curious whether it will be accurate enough for our purposes. It would certainly be much more time consuming. Yes, resolution of product length (esp fully extended) is the goal.
CJ (10-30): This is something we need to try by experiments. If the gel does not work, we may consider EC, which is claimed to be more sensitive and accurate.
d) Ideally, because the rate of nucleotide addition for the last dNTP added is different than that of all other dNTPs, it is good to use a long template, especially if we are making predictions of total fluorescence at later times. However, since we did not consider this to be an issue for MM kinetics, we will ignore it here as well and it is not a priority to work with a new template for robustness analysis.
I agree. A long ssDNA may be easier to form 2nd structure. Also, chemical synthesis of oligos >90nt is not quite practical. Some companies claim they can synthesize oligos of ~100nt, but I would be very cautious with the quality of their products.
RC 10/28/2013
Thanks for the revised draft, I've looked over it and have some comments on the revisions as well as the next step for experiments (should we choose to proceed with them at this time):
-- Manuscript. Please comment on the following:
a) What was the fate of Sudha's work on Km determination for the DNA? Did you eliminate this because of the protocol she used? If so, what were the issues? Was the reason that a lot of the original manuscript was deleted?
b) Where (what page) did you include the discussion about nucleotide inhibition?
c) The robustness analysis (if included) will be done using the method different from (and perhaps shorter than) that you have included in the draft. I will revise that section. At the same time, I will decide on the content of the simulation section. How much of this we choose to include will affect the choice of journal.
d) What are some example journals you referring to when you indicate a 7000 word limit?
e) You did not appear to consider an Arrhenius model for extension rates as a function of temperature (i.e., a figure that examines whether a constant preexponential factor and activation energy can accurately predict the temperature variation of extension rates). Is this something you plan to add?
-- Robustness experiments. I've read your comments below. Here are some further details on how the experiments used for model validation (as opposed to MM parameter fitting) may differ. This list is in order of priority. Bear in mind that we may not do include the results of all such experiments for this paper (we may put them in a second paper instead).
a) We need to quantify the standard error of measurements of total DNA concentration for a known total concentration of DNA. This is because the difference between the model predicted total DNA concentration, given an estimated value of kcat/Kn, and the measured total DNA concentration is attributable to both the error in the model prediction (due to e.g. error in the kcat/Kn estimate) and the measurement error, and we are interested in the former.
b) Regarding the protocol suggested below, it is good to see that the conditions are similar to those in PCR. Please indicate how your lowest [dNTP] compare to those used in the later cycles of PCR (where nucleotide gets depleted). What are some characteristic values for the latter? The model can be used to predict incorporated [dNTP] under non pseudo-first order conditions as well, but we should first verify whether those are relevant.
Did you find that the first-order kinetic model fit the data equally well at your lowest vs highest [dNTP], indicating that a pseudo-first order approximation was appropriate at all [dNTP] concentrations?
While we may not change the protocol given that it suits PCR, we may want take more measurements of total DNA concentration at particular times to get a better estimate of the experimental uncertainty when comparing to model predictions.
c) Though the current measurements can give us the total concentration of incorporated nucleotides, in order to determine the total concentration of fully extended DNA at a particular time (which the theoretical model can predict and which is of greatest interest in PCR), we may need to run a gel and extract the fully extended product. Please provide comments on how difficult this would be and the associated measurement error of the fully extended DNA concentration (esp how it compares to the fluorescence measurement error of the total incorporated dNTP concentration).
d) Ideally, because the rate of nucleotide addition for the last dNTP added is different than that of all other dNTPs, it is good to use a long template, especially if we are making predictions of total fluorescence at later times. However, since we did not consider this to be an issue for MM kinetics, we will ignore it here as well and it is not a priority to work with a new template for robustness analysis.
CJ 10/25/2013
Attached please find the extension kinetics manuscript, revised based on our new plan and new data. Notably, most journals have a word limit of <7000 words for articles. The previous version of manuscript has >9000 words, and we need to add a lot of work on simulation. Therefore I trimmed the background and discussion section, making them more compact and concise. I have left comments in the file to indicate where to fill in the theoretical work. I am also attaching a track change version in case you want to know what changes did I make to the old manuscript.
Taq Paper CJ 102513.doc
Taq Paper CJ 102513 track change.doc
Raj, regarding your comment, I would like to get clarified on what difference we want to make in the next step. You want to measure [total DNA (and partially extended primer-template) concentration as a function of time starting with the addition of nucleotide to pre-annealed and enzyme-bound primer-template (see below), under conditions of significant nucleotide excess as is common in the initial cycles of PCR (in PCR extension the enzyme and primer are also in excess in the initial cycles)] Actually this is exactly what we have been doing: we measure dsDNA extension as a function of time; we use pre-annealed and enzyme-bound primer-template; and we use excessive dNTPs (eg. 200 - 1000 uM) for some groups of experiments. You also mentioned [The protocol would differ from the MM kinetics protocol in that we are no longer interested in just the initial rate.] As a matter of fact, in many groups (high dNTP, high temperature) of our previous expts, the reaction has reached plateau, not just in in the initial stage (see a representative figure below). I also want to mention that conditions in our previous experiment, in terms of template and enzyme concentration, Mg concentration, dNTP concentration etc., are quite comparable to what in real world PCR application. The only significant difference is the extension time: usually it is 1min for 1kb, but we do 10min for 80bp. Overall, I am not quite clear on what kind of protocol you want me to design for the next step. Would you possibly clarify on what is the purpose for this project and how it is different from the previous one?
The figure below is a representative reaction curve that proceeded well beyond the initial stage, measure at 65 C, 1000uM dNTP.

RC (10-23): Thanks. After the completion of MM extension experiments and experimental manuscript section, the knowledge transfer on the b-lactamase project, and the literature review, etc on the diagnostics projects, you should consider preparation of a protocol for measurement of total DNA (and partially extended primer-template) concentration as a function of time starting with the addition of nucleotide to pre-annealed and enzyme-bound primer-template (see below), under conditions of significant nucleotide excess as is common in the initial cycles of PCR (in PCR extension the enzyme and primer are also in excess in the initial cycles). The protocol would differ from the MM kinetics protocol in that we are no longer interested in just the initial rate. However, as in the MM protocol, we do not want to consider the time course of primer annealing and enzyme binding to primer-template hybrid. We would like to record the total fluorescence and its standard error at regular sampling intervals. These results would probably not go in the current manuscript, but we may write a follow up manuscript that uses them.
CJ 10/21/2013
Attached please see a summary of the extension experiments I've been running so far. These data will be integrated into the extension kinetics manuscript, which I'm currently working on.
102113 CJ report.ppt
RC (10-2): Thanks for the recent updates. As mentioned below, we can keep revising the experimental parts of the extension paper draft in the meantime.
I have worked on the theory parts of the extension paper including the robustness analysis. I will discuss with you after the above are further along.
We will also have to come to a judgment regarding whether we want to run any extension reactions under PCR conditions in order to check the predictions/robustness
of the model based on the MM parameters. This would require a new type of experiment that does not just measure initial rates but monitors fluorescence during the whole extension reaction - which we may not want to include in this paper. We should come to a conclusion on this because it affects how we will present the modeling and robustness analysis parts of the paper (they may be made shorter if we want to address the later issues in another paper). This will be easier to do after seeing the layout of the rest of the paper.
CJ 9/20/13
Attached please find a report on the experiments I ran this week (60C).
092013 CJ report.ppt
CJ 9/13/13
Attached please find a report on the experiments I ran this week (50C).
091313 CJ report.ppt
CJ 9/6/2013
Attached please find a report on the experiments I ran this week (55C).
090613 CJ report.ppt
CJ 9/5/2013
Attached please find an initial framework of the Taq extension kinetics paper, along with my comments. Please let me know how it should be revised. After figuring out the general structure of the paper I will start with the write-up.
Currently I'm also running the experiments for this paper, which would take ~4 weeks. Hopefully the simulation work and robustness analysis could be done during this time period.
Taq Paper CJ 090413.doc
RC: I will comment shortly. No further theoretical write up, simulation work or robustness analysis should proceed until then. Experimental parts can continue to be revised.
CJ 9/4/2013
Meeting with Karthik:
Plan for the extension paper:
(1) CJ will write a framework of the paper by the end of this week based on Sudha's previous manuscript.
(2) Karthik will post some references for the robustness analysis. CJ will read them first; then discuss with Karthik if there's any question.
RC: RC will provide needed references since there is work underway in the group on this.
(3) CJ and Karthik will then fill out the paper with expt and simulation results.
CJ 9/3/2013
Thanks Raj! I will work on the write up as soon as I get an electronic copy of Sudha's manuscript. I went through all the files uploaded on Wiki ever, but did not see that one. Karthik, would you kindly help me to find that file and send it to me?
Raj sent me a version in October and I don't know Sudha has updated that draft after this. Here is a draft that I have.
Thanks Karthik! This is exactly what I'm looking for. - CJ
Taq Paper Draft April 10-1.doc
RC 9/3/2013
KM, I have looked over the extension robustness outline. Thanks. Once we have settled the simulation plans, I will revise this as necessary, and also will revise/extend the BP paper robustness analysis, which was also based in part on Nagy's papers. As you know, in our group Andy has worked extensively on robustness analysis, including simulations using these approaches, and you should communicate with him regarding his experiences. The theory outlined below has been discussed and implemented in Andy's control work (see e.g. our slides on robustness analysis of qc and his current working papers). The Taylor approximation for the computational robustness analysis is useful primarily in an optimization context due to its speed. However, it is often inaccurate; Andy can give you details. We have developed methods in our group that are significantly more accurate for time-varying linear systems (stage 1 of PCR). I will comment on this, but before I can do this we need to settle some details of the scope of what we will be presenting in this paper. The presentation should be brief.
What I would like you to work on now is the detailed plan for how the simulations will be compared to the experiments:
- For this paper, there is no reason not to obtain the state variable uncertainties through simulation, since it will be more accurate. We should start setting up the code for this. It may be useful to talk to AK about it.
KM: Yes, we should obtain the state variables uncertainties through simulation and it was actually the original plan. It can be done in many ways.
1) By sampling the worst case parameters and solve the state equations.
2) Robustness analysis will give the distribution for the evolution of the state variables and it should talk about the uncertainty in the state variables.
Do you mean 1) when say that obtain the state variable uncertainties through simulation.
- CJ will be providing experimental uncertainties for the kcat/Kn parameter. Per our previous discussions, I believe this was to be the only uncertain parameter in our extension model, since we will be starting with fully bound enzyme. CJ and KM, please confirm. We should start by sampling from the normal distribution of this parameter value and computing the moments of the state variable.
Yes, we need only kcat/Kn parameter.
- We also need to decide on what will be the state variable of interest. I believe we agree that this should be the sum of concentrations of all E.Di's, since this is what can be measured experimentally. CJ and KM please confirm.
Yes,
- We need to decide on what experimental conditions will be used for this analysis. Are we planning to use current assay conditions, or PCR conditions? Are the conditions similar? How do they differ?
Ideally a rate constant is only a function of temperature. To start with we can use the current assay conditions but I think it would be appropriate to use the PCR condition for the comparison with the theoretical results. CJ can comment on the conditions.
- Are the assay conditions suitable for application of pseudo first-order kinetics (i.e., stage 1 of PCR)?
As stated above as long as we have a rate constant which is a function of temperature , it should be sufficient and we should not worry about assay conditions.
- For the theory part, are we planning to consider variable extension temperature or only constant extension temperature? The AK/RC method is most useful for variable extension temperature with pseudo first-order kinetics. It is more important to mention it in that case.
In the current plan, I have included the variable temperature. This is important, especially for the simultaneous annealing and extension.
CJ, please start by putting together all the materials we have so far regarding the experimental estimation of the MM (kcat/Kn) parameter, and whether/how we will be using Sudha's earlier work
on Km estimation.
This should be connected with the materials KM and I put together on the MM theory.
Then later you can add the theory on Mg chelation.
KM, please set a time to discuss these plans w CJ.
I will meet CJ on Thursday. Will send an email to CJ regarding this.
CJ 9/3/2013
Thanks Karthik.
"by fixing E.SP complex and Nucletide (dNTP) concentration (Need to make sure there is no SP) conduct an extension reaction at a fixed reaction temperature (may be 72 deg C). During the course of the reaction, measure the concentration of DNA (completely extended DNA or whatever the variable that can be measured with respect to time)."
This is exactly what I'm doing right now. By running such measurements I get reaction curve like this:

Is this what you are looking for? Please be advised that we are actually not monitoring the extension in a real-time manner. The real-time PCR, according to Sudha, does not work quite well in our experimental setting (never tried myself). So we run the extension for various period of time; quench the reaction, and then quantify the amount of dsDNA. Please let me whether such kind of data would be useful for your robustness assessment. If not, please let me know how you want them to be improved.
CJ, as far as the experimental validation, this should be sufficient. We can theoretically produce this kind of plot by solving the extension state equation and compare. (RFU can be converted in to equivalent concentration.
CJ 9/3/2013
A quick question for Karthik:
In the attachment you posted today, you suggested a protocol to experimentally verify the robustness of the model parameters (on the last page):
1.At a fixed reaction condition, experimentally estimate the evolution of a specific state variables (DNA concentration). This can be done in the PCR machine.
So right now we have following variables in our PCR reaction: (1) Taq concentration; (2) template concentration; (3) dNTP concentration; (4) Mg concentration; and (5) extension temperature. By saying 'a fixed reaction condition', you mean fixing which ones of (1)-(5)? I suppose by saying "DNA concentration", you actually means template concentration. Then what do you mean by 'estimate the evolution of a specific state variables (DNA concentration)'? Would you kindly elaborate your general experimental design?
KM(9/3/2013)
What I meant was, by fixing E.SP complex and Nucletide (dNTP) concentration (Need to make sure there is no SP) conduct an extension reaction at a fixed reaction temperature (may be 72 deg C). During the course of the reaction, measure the concentration of DNA (completely extended DNA or whatever the variable that can be measured with respect to time). Mg and dNTP concentration can also be fixed.
For the fixed values of the above concentrations, we can solve the model and check whether the model is robust or not, by comparing the theoretical and experimental results. Hence, we can comment on the robustness of the model.
KM 9/3/2013
Outline and plan for the robustness analysis of the extension model.
Robustness Analysis of Extension Reaction.docx
Karthik.
CJ8/30/2013
Attached please find two references: (1) a comprehensive review on the stability constant of Mg-ATP complex (much more data than the paper I showed Raj). (2) the BioTechniques paper the Mg calculator cited. I also made a spreadsheet to calculate free Mg concentration, also attached. My results are slightly different from the results given by the online Mg calculator (maybe due to ion strength? I'm still inspecting). But the bottom line is, when [dNTP] ranges from 0.2 - 1 mM, the formation of Mg-dNTP complex is near-stoichiochemical. We can roughly estimate free Mg concentration by the following formula: [Mg]free ~ [Mg]total - [dNTP].
The first reference includes stability constants of various NTP with Mg (Table 6). We can see that different bases show little effect on Ka. The temperature does not seem to strongly affect the stability constant either (Table 3).
As for the difference between NTP and dNTP, I found that ATP chelates Mg by its phosphate groups. The OH groups on the ribose ring are not directly involved. Therefore the stability constant of dNTP-Mg is expected to be close to that of NTP-Mg.
In conclusion, we can adjust the Mg concentration based on the formula [Mg]free ~ [Mg]total - [dNTP], keeping [Mg]free constant as varying [dNTP]. Next week I'm running one set of PCR using the following condition:
2nM Taq; 200nM template; 55C extension temperature; [dNTP] = 2, 10, 50, 100, 200, 400, 600, 1000uM. The [Mg] will be adjusted so that free [Mg] is maintained at 2mM. Based on my preliminary results (8/29/2013), I anticipate we would not see the inhibition at high dNTP concentration any more.
free Mg calculator.xls
CJ 8/30/2013
I found an online calculator that can calculate free Mg concentration based on Mg-ATP complex formation:
http://maxchelator.stanford.edu/MgATP-TS.htm
The dNTP may have slightly different stability constant compared to ATP, but the difference should not be very significant.
CJ 8/29/13
Please see a report on my recent experiments attached:
- We now have two strategies to deal with the Mg chelation issue, both seemed to work in preliminary test:
(B) Run PCR with varying MgCl2, keeping the free [Mg2+] constantly at 2mM.
- My concerns about method (A) are: (1) My preliminary results on Mg dependence of Taq somehow does not agree with literature reports. I need to re-confirm it under various conditions (varying [dNTP], temperature etc). (2) 10mM MgCl2 is not recommended for real PCR applications, probably due to fidelity issue. (3) The reaction may be too fast to sample at higher temperature and [dNTP]. But I can fine-tune it by adjusting Taq concentration.
- My concerns about method (B) is that the real concentration of free Mg2+ is not known, and currently hard to calculate. One possible solution is to measure the Kd of dNTP binding Mg2+ by ITC (isothermal titration calorimetry). I know Columbia has one such instrument open for public use with fair hourly rate. Other universities such like Princeton, Purdue, or CMU may also have similar service.
- I personally prefer method (B). I will keep on searching for literature values of Kd between dNTP and Mg2+.
- Meanwhile I am curious about the Mg dependence of Taq activity. I would like to run a series of PCR at 75C with [MgCl2] varying from 1mM to 10mM. However, this is not at high priority. I would like to hear Raj and Karthik’s feedbacks before setting up the expt.
I also want to confirm with Raj and Karthik that I'm going to repeat the entire set of experiment Sudha did, with 2nM Taq instead (I will figure out the Mg concentration before running the large scale measurements). For this I will order 5000U Taq from Invitrogen, costing ~$750 after discount.
082913 CJ report.ppt
CJ 8/26/13
Please see attached the report on my recent experiments.
Regarding the Mg chelating issue, I’m thinking of two possible solutions:
–Adjust Mg concentration so that free Mg is kept constantly at 2mM. To do this I need the equilibrium constant of dNTP chelating Mg. I’m still searching for literature values of this constant, or a straightforward way to measure it.
–Or, we run the reaction at a higher Mg concentration, so that even with 1000uM dNTP, the chelating would not be a significant interference. To do this we first need to make sure that Taq is happy with 10mM Mg. I would like to justify this in the next round of expt.
082613 CJ report.ppt
SM 8/23/13:
Please see following files for a summary on the role/ interaction of [Mg] and [dNTP] on the activity and fidelity of various DNA polymerases. The excel file summarizes these publications in terms of relevant protocols, reaction compositions, observations and data and also includes pointers to specific tables or fig.s in corresponding papers. The summary is not exhaustive at this point. Probably I will add more papers as well as more details in the spread sheet.
Literature summary 081913.xlsx
NEB Guidelines for PCR optimisation.pdf
Kinetic Analysis of Escherichia Coli Deoxyribonucleic Acid PolI J. Biol. Chem.-1975-Travaglini-8647-56.pdf
Fidelity of Therrmococcus litoralis (Vent) DNA polymerase.pdf
Fidelity of DNA synthesis by the Therrmococcus litoralis (Vent) DNA Polymerase.pdf
Fidelity of DNA Synthesis by the Thermus aquaticus DNA Polymerase.pdf
Elementary Steps in the DNA Polymerase I Reaction Pathway.pdf
Optimization of the PCR with regard to fidelity.pdf
Isolation, Characterization, and Expression in Escherichia coli of the Taq DNA polymerase.pdf
High-level expression, purification, and enzymatic characterization of taq polymerase.pdf
High fidelity DNA synthesis by the Therrmus aquaticus.pdf
DNA sequencing with Thermus aquaticus DNA polymerase.pdf
DNA Polymerase Insertion Fidelity.pdf
Analysis of DNA polymerase activity in vitro using non-radioactive primer extension assay.pdf
An Induced-Fit Kinetic Mechanism for DNA Replication Fidelity Direct Measurement by single turnover kinetics.pdf
A Biochemical Perspective of the Polymerase Chain Reaction.pdf
Pre-Steady-State Kinetic Analysis of Processive DNA Replication.pdf
Kinetic Mechanism of DNA Polymerase I (Klenow).pdf
Kinetic Analysis of Escherichia Coli Deoxyribonucleic Acid PolI J. Biol. Chem.-1975-Travaglini-8647-56.pdf
RC (8-22): Sudha, have you posted the literature on inhibition?
CJ (8/19/2013)
Thanks for the reference from Karthik! Notably, in that paper the [KCl]=75mM, still higher than what we use (~ 15mM).
Regarding Raj's question "what point you will be able to determine whether your data displays inhibition kinetics." As I have said in the 8/19 report, my next expt would be under high dNTP concentration. Currently we run out of Taq enzymes. Sudha and I have ordered more and I will run the experiments once they arrive (later this week).
KM(8/19)
Datta and Licata used the Taq polymerase.
Thermodynamics of the binding of Taq DNA polymerase to primer-template DNA.pdf
CJ (8/19)
Regarding Raj's comments and questions on 08/15/13:
RC: a) How this will be presented: I believe the Kd you have used below is based on Km from Sudha's original experiments. I assume we will be commenting on the uncertainty in the calculation of E.SP based on this Kd and presenting the 200 nM and 400 nM results in order to demonstrate that the enzyme is mostly saturated at 200 nM? Have we concluded that the Data and LiCata reaction conditions were different from those we are using and hence we cannot use that Keq?
CJ: I'm using the Km from Sudha's original expt (~10nM) as reference to calculate E.SP. In the Datta and LiCata paper, first of all, they used Klentaq (5' nuclease domain removed) instead of taq; and their KCl concentrations (50 - 500mM) were much higher than what we use (~15mM, from taq buffer and the enzyme solution). Their Mg concentraion (5 mM) was also different from what we use (2 mM).
If you look at Fig.4 on Datta and LiCata's paper, what we can do is extrapolating the line to 5mM (ln[KCl] ~ -5). In that case the y value should be well above 18, which means the Kd is lower than 15nM. (ln 1/Kd = 18 means Kd ~ 15 nM). Therefore if we use the Datta and LiCata's results as reference, we can still conclude that the Kd of taq binding to template should be at low nM range.
RC: b) Do we expect the higher enzyme concentration to also help reduce the degree of any inhibition by nucleotide, since I assume we will be using the same concentrations of nucleotide?
CJ: I'm not sure about this. Actually, I would like to try high dNTP concentration in the next expt.
RC: c) I agree with the proposed next set of experiments
CJ: Please see a report attached.
081913 CJ report.ppt
RC (8/19):
I agree that chelation of Mg by dNTP should be looked into. As noted, we need to be careful before attributing any
inhibition effect to binding of dNTP to the enzyme. This could be a more plausible explanation for inhibition since we have a physical picture
for how it occurs.
1) Can you post some of the literature you mention below regarding what is known regarding chelation to the wiki?
2) [dNTP] concentration varies across the assays so any chelated [Mg] will vary. As in the case of excess template concentration, if we choose
to increase the Mg concentration we would need to determine the appropriate concentration. Without looking at the literature I do not
know how high these [Mg] concentrations would need to be. Note that if the [Mg] bound to enzyme increases above standard concentrations in PCR,
it could be a problem. Are you currently using [Mg] similar to that used in PCR?
3) In the meantime, I suggest we continue with the assays Chaoran is carrying out to improve the signal to noise. We will see whether the
inhibition effect is observed in these assays as well despite the reduction of noise. Then we can decide on next steps.
Chaoran, please let me know at what point you will be able to determine whether your data displays inhibition kinetics.
SM (8/15):
As a follow up of our meeting last week (8/9/13), I have done a detailed
literature search on substrate inhibition by dNTPs as seen in my assays.
The following is the summary:
1. Websites and technical literature of most polymerase
manufacturers (eg. NEB, BioRad etc) state that 200uM dNTP is optimum for
most polymerases and that excess dNTPs can chelate Mg2+ and thus inhibit
the polymerase. This may be avoided by keeping [Mg] well in excess of
[dNTP]. However, the original published paper (if any) for this
statement has not been cited anywhere.
2. There are lots of publications about [dNTP] and [Mg] and
fidelity of the polymerase. However, these do not talk about the
kinetics.
3. Only 1 publication (Huang, Norman and Goodman, Nucleic Acids
Research, Vol. 20, No. 17 4567-4573) mentions Michaelis-Menten kinetics
with regards to varying [dNTP]: In the methods section, pg 4568 (Even
in this publication though, there is no data to support this statement):
"The primer elongation velocity is defined as the percent primer
extension/minute. Plotting v versus [dNTP] fits a Michaelis-Menten
equation, and the apparent second order rate constant, Vmax/Km for each
primer terminus was determined by non-linear least squares fit to a
Michaelis-Menten curve."
...
RC (8/15):
Regarding updates below:
a) How this will be presented: I believe the Kd you have used below is based on Km from Sudha's original experiments. I assume we will be commenting on the uncertainty in the calculation of E.SP based on this Kd and presenting the 200 nM and 400 nM results in order to demonstrate that the enzyme is mostly saturated at 200 nM? Have we concluded that the Data and LiCata reaction conditions were different from those we are using and hence we cannot use that Keq?
b) Do we expect the higher enzyme concentration to also help reduce the degree of any inhibition by nucleotide, since I assume we will be using the same concentrations of nucleotide?
c) I agree with the proposed next set of experiments
KM(8/14)
I was working with the fixed enzyme concentration of 0.36 nM (Because this was the limit that worked well before and based on this I calculated the template concentration) of enzyme concentration and found the suitable template concentration and that is how I ended up with 200 nM. (As you can see, though 20 nM is higher than the Kd value it does not give more than 66% of equilibrium conversion).
I have checked my calculation with your values and it is correct. For 2nM of enzyme and 200 nM of SP, it is possible to get 95% conversion.
CJ (8/14):
Please see my progress report attached.
Karthik, I have different opinion on the enzyme concentration. As I have explained, if we have template concentration much higher than Kd, we will have almost all enzymes binding to template, regardless of the enzyme concentration (within a certain range, of course). Please see a calculation in my progress report. Basically, using 2nM taq with 200nM template would result in 95% of enzyme binding to template. And if you use 0.36nM taq with 200nM template, although the enzyme concentration is much lower, the % of enzyme binding to template remains mostly the same. (I was assuming a 10nM Kd of taq binding to template). I'm also attaching a spread sheet to calculate the % enzyme binding to template. You may play with it if you like.
081413 report - wiki.ppt
E.SP calculator.xls
KM (8/12):
Reason to keep low enzyme concentration
1. For a fixed template concentration, a low concentration of the
enzyme ensures that all the enzyme molecules binds on the template.
2. It is important for all the enzyme molecules to bind on the
template otherwise
a. It will be difficult to measure the sum of E.D_i molecules.
Because, if there is any free enzyme in the reaction at the start of the
reaction, as soon as the extension reaction starts, these free enzymes can
react with template to produce more E.D_i molecules. We don't know how many
such enzyme molecules reacted with template to produce more E.Di. In this
way, we can't make any approximation that initial concentration of the
Enzyme is equal to the summation of all E.Di molecules.
b. It is important to measure the summation of E.Di molecules for the
MM kinetics formulation.
SM (8/12/13): Minutes of meeting 8/9/13
1. Curve fitting of Rate Vs [substrate] according to a substrate inhibition model was discussed:
Activity Assays for Taq Polymerase at 50-75oC with dNTP concentration variation 2-500uM repeatedly had the following observation: increasing Taq activity with increasing [dNTP] upto 200uM, with further increase in [dNTP], there is a decrease in Taq activity.
In the initial data analysis, the traditional Michaelis-Menten equation was used for curve fitting, however, it was seen that the fit was very poor.
Substrate Inhibition model was tried as an alternative after KM and SM’s meeting (8/5/13). This model gives good fits for the existing data. No manual removal of outliers has been necessary.
Prism calculates Vmax, Km and Ki values for assay temperatures (55, 60, 65oC) but is unable to calculate for the remaining temperatures (50, 70, 75oC). Taq Activity may be too low at 50oC for the curve to be reliable and at 70oC, we did not have enough data points. However, KM feels that these are good fits, and it would be possible to calculate Vmax and Km ourselves.
Literature search to support the experimental results is ongoing and will be summarized in a separate post.
2. Improvement of the experimental noise was discussed:
The issues that need to be considered are:
a) getting accurate RFU values for early time points,
b) reducing background as much as possible, and
c) reducing standard error in RFU values.
d) getting a significant net gain in RFU for the complete incubation.
- The experimental protocol as standardized to date cannot be improved further with the current experimental set up limitations. In fact, comparison of corresponding assay results from Jan 2012 and July 2013 shows identical activities for Taq polymerase showing that the current protocol works consistently.
Under the given assay conditions of template, Taq, dNTP concentrations, the rate vs [dNTP] curves may be improved only by increasing the number of data points (assays at more dNTP concentrations eg., 50uM, 150uM, 250uM and probably 1000uM; currently data exists for 2, 10, 20, 100, 200, 300, 400 and 500uM dNTPs). However, the entire series would again have to be assayed (the complete dNTP range over the complete temperature range using the same batch of reaction components) for the activities to fit in the same rate vs [dNTP] curves. The feasibility of doing the complete series needs to be determined in terms of time (this can again take up to 3-4 weeks) and material (some reagents will need to be ordered).
- CJ has suggested exploring different assay conditions for reduction of noise. This will be useful information to improve experimental results. The approach to date for the current series of experiments (Apr-July 2013: dNTP variation) has been to experimentally validate the calculated template: enzyme ratio. This has led to issues of noise that cannot be improved due to experimental limitations (see point above); in the new approach, the idea is to get the experimentally optimum curves and then calculate back to the percentage of template-bound enzyme and thus, the template: enzyme ratio.
In this context, it will good to note assay conditions tested to date and comments on results obtained. Please see attached table for summary.
3. Review of the original manuscript needs to be done to determine if the Vmax, Km calculations can be related to current calculations.
Karthik, Chaoran, please feel free to edit/ add.
Sudha.
Taq Pol Assay Conditions
SM 8/12/13:
I will post minutes as well as the experimental parameters that have been tried so far.
RC (8/9/13):
I would appreciate it if someone from PMC-AT would post the minutes of the meeting today and if Sudha could post the parameters in the experimental set up
that introduce noise.
Broadly speaking it appears the inhibition model and associated experiments will be finished while exploring conditions for the reduction of noise. The latter will also verify whether inhibition occurs for other experimental conditions as well. For example, if enzyme concentration is increased, do we still expect to see strong inhibition?
CJ 8/9/13
Here is a paper that has a brief description on the substrate inhibition model. The equation 1 in this paper is the exact equation Sudha used to fit the curves in her recent report.
http://onlinelibrary.wiley.com/doi/10.1002/bies.200900167/pdf
SM 8/8/13:
Karthik has shared the excel file generated by him where he has manually removed outliers in part of the 55oC experimental data. Sudha will attempt Prism curve fitting with this subset data to determine if curves improve.
- They do not improve.
- The Prism software can itself detect outliers. It might be better to use the Prism generated information as a basis and then improve (?) on it manually.
- The 6 replicates (sum of 2 experiments) cannot be taken as 6 individual replicates due to batch-to-batch variations.
A possibly better fit than “one-phase association” kinetics may also be investigated since most time courses in the present series of experiments show a linear increase in RFU with time even up to 10 mins (the plateauing typical of one-phase kinetics is not seen).
- See attached file: review and comparison of older experiments with current data. One-phase association kinetics need not be ruled out completely.
2. Given the observation that initial reaction rate increases with increasing [dNTP] up to a point and further decreases with increasing [dNTP], the appropriateness of using the Michaelis-Menten equations for fitting Initial Rate vs Substrate Concentration was discussed.
- See attached file: curve fitting using equation for substrate inhibition. This equation still allows us to calculate Vmax and Km and also gives Ki.
Karthik and Sudha will both be doing a literature review to determine if polymerase kinetics with varying [dNTP] has been discussed previously.
This is ongoing.
Sudha.suppl to taq pol assay 50-75deg aug 2013
RC (8/7/13):
Some feedback regarding the history of this issue vis-a-vis the current work (to the
best of my recollection, since we had these discussions several months ago):
1) We considered using Sudha's Km data from the original manuscript in
order to do calculations of the amount of template bound to the enzyme
(i.e., the "[E.Di]_0" in the MM terminology). This way we could make use
of the original data in the manuscript. We also found literature data
regarding the enzyme-template equilibrium constant at various
temperatures. I don't recall offhand if this Keq was for Taq. If you
can't find this literature data on the wiki, let me know - I think
Karthik may not have posted it.
2) I believe that in order to avoid propagating uncertainties from Km or
Keq at each temperature, we initially opted for a strategy wherein we
would use saturating concentrations of the template. At this point we
were not aware of the experimental noise. As described in protocol pt 7
of "extension paper theoretical part", whether we had in fact used
saturating concentrations could be checked by increasing the template
concentration slightly further and checking for a change of
fluorescence. This check did not require accurate knowledge of Km at the
given temperature.
3) Now that we know that we are encountering issues with noise, I agree
that we may need to use an approach where we rely on a calculation of
[E.Di]_0 rather than operating under saturating conditions.
4) One could consider using either Km or Keq to calculate [E.Di]_0. Note
that this extension reaction has some differences with respect to other
enzymatic reactions where MM kinetics is applied because the catalytic
step of nucleotide addition does not change [E.Di] unless the last
nucleotide is being added. Thus use of Km for template binding to
enzyme, which has a contribution from kcat, may not provide an accurate
estimate of [E.Di]_0. We may have opted to use Keq in order to avoid
such issues (as far as I can recall).
5) I would suggest proceeding as you suggest, using the Km from Sudha's
initial draft to calculate [E.Di]_0, and then comparing to the results
using Keq if you can get Keq. The difference will depend on the magnitude of kcat.
6) We should bear in mind the experimental protocol, wherein the enzyme is incubated with
template for several minutes prior to adding nucleotide. This provides sufficient time for the equilibrium
value of [E.Di]_0 to be achieved.
7) If we do proceed by using Sudha's estimates of Km (Michaelis constant
with respect to template) it would be a nice way to connect up the old
version of the manuscript with the new. In that case, we would also
revise the "extension paper theoretical part" to indicate that [E.Di]_0
is calculated using Km (template) or Keq.
8) I am not sure if we will need the iteration methods you mention
below, but we can discuss those issues soon for clarification.
If we rely on Km as estimated in the original manuscript as the basis for calculation
of the kcat,Kn for nucleotide addition, I would like us to review that part of the manuscript
at least once to make sure Km in the manuscript is defined in the same way we are
assuming.
If I recall any other aspects of the history of this issue (or if I need to edit the above), I will
do so in another update.
CJ (8/6/13):
I'm calculating the %ES (percentage of taq in bound state)
based on the Km in Sudha's original manuscript. Then I will get an
estimation on what would be the template:taq ratio to start with.
Another thing in my mind: if there's really a dilemma between the theoretically desired template:taq ratio versus
what experimentally desired value to get good signal-to-background, I'm thinking of one
thing we can do: experimentally we use a lower template:taq ratio, say 50 - 100 instead of 1000. Then during
data processing, instead of assuming all taq is in bound state, we actually calculate the percentage
of enzymes in bound state. This would just be a quadratic formula. The only issue is that we should know the Km.
Of course we don't know it before fitting the curve. However, I think we can first make a guess
based on preliminary data, and then do iteration until the calculated Km converges with the guessed number.
(Sorry I'm not quite good at the math terms, please let me know if I failed to make it clear.) In this way the
data processing will be a little bit more complicated. But this would be the back-up plan if we can not get acceptable
results from the current setup.
So this is what I will do in the next couple of days:
- I will repeat one group of Sudha's experiment, just as training for me to get familiar with all the instruments. The condition will be: 0.036 nM taq; 20 nM template; 200 uM dNTP; 70C.
- I will calculate the %ES and try to fine tune the taq and templat concentration in order to improve the quality of data. Will come up with a proposal by Wed or so.
- If we would like to try the iteration methods, I will think of it and start to work on Matlab, if we do have access to it.
RC (8/6/13):
There were a couple of questions raised regarding the protocol.
1) This reaction involves 2 steps - a) enzyme binding to single
strand-primer hybrid and b) nucleotide binding to the E.SP complex
followed by bond formation.
2) In the new experiments we are applying a MM model to estimate Kn,
kcat for b)
3) In b), the "enzyme" is really E.SP (or E.Di) hybrid. It is not E.
4) So that we can approximate the concentration of E.SP hybrid (the
"enzyme" concentration), we keep the SP concentration high so we can
assume nearly all the enzyme is in E.SP form. Then "E.SP_0" can be taken to be approximately equal to
E_0. To get an idea for how high it should be, we use a known
approximate value for the equilibrium constant for E.SP binding from the
literature. I believe this is how Karthik chose the SP concentration,
and I believe this is discussed on the wiki.
5) In order for MM kinetics to hold for b), we need to keep nucleotide
concentration high. This justifies the steady state assumption of MM
kinetics for b).
8/6/13
Minutes: 8/5/13 meeting
1. Karthik has shared the excel file generated by him where he has manually removed outliers in part of the 55oC experimental data. Sudha will attempt Prism curve fitting with this subset data to determine if curves improve. A possibly better fit than “one-phase association” kinetics may also be investigated since most time courses in the present series of experiments show a linear increase in RFU with time even up to 10 mins (the plateauing typical of one-phase kinetics is not seen). The feasibility of manually removing outliers from all the data sets needs to be determined.
2. Given the observation that initial reaction rate increases with increasing [dNTP] up to a point and further decreases with increasing [dNTP], the appropriateness of using the Michaelis-Menten equations for fitting Initial Rate vs Substrate Concentration was discussed. Karthik and Sudha will both be doing a literature review to determine if polymerase kinetics with varying [dNTP] has been discussed previously.
3. Given the limitations with experimental setup, the variability may/ may not be reduced further by altering test conditions (eg., template concentrations etc). However, such alterations should not compromise significance of net RFU gain during the time course. Repetition of experiments will be attempted only after data analysis options have been explored.
(Karthik: The file you sent me has only data for 2, 20, 100uM dNTPs, is this the 55deg expt? Also do you have excel files for the other temp.s?Please feel free to edit the minutes.)
Sudha.
RC (7-29): Do you mean that you and Sudha have used different methods to compute the initial rates and that the results do not agree, or has only Karthik calculated initial rates? I assume you are suggesting some of these experiments need to be repeated?
KM(7-29). Raj, the initial rate that was provided by Sudha is based the Prism fitting which was not proper (as per R^2 value of the plot). So we decided to go through each data manually ( please read the minutes of our meeting) and remove some of them (essentially we have 6 replicates). I did that but I could not improve the quality of plot. So we need to discuss on removing some data (we need to identify a proper way) and redo the calculations using the Prism. Decision about repeating some experiments can be taken once we finalize the data analysis part.
Karthik.
7/29/2013
Sudha,I have gone through the data and tried to analyze based on the following
- I tried to remove few data based on the following strategy
- For a fixed substrate concentration and a temperature there are 12 time and concentration data.
- There are 2 trials for each data with 3 replicates for each trial.
- Therefore we will have 6 data points for each time interval.
- I found the average based on the each data point.
- Then manually deleted 2-3 points that deviates too much from the mean and reduced the variability.
- The DNA concentration at time, t = 0, has been calculated as per the above approach and based on -taq value. Note that here will have 18 data points ( 3 different time interval).
- Once I found the average RFU values at all the time intervals, I removed 4 to 5 time interval that does not follow the theoretical trend of DNA concentration. For example, ideally, from time, t = 0, the DNA concentration should increase and reach the saturation value. But there are few data violated this observation and I removed those data.
- Finally I calculated the initial rate based on numerical derivative.
- As per the above method, the initial rates that I have calculated were not consistent. Between 3 time intervals it varied too much or I could not make a small discretization to calculate the initial rate because I did not fit the data.
- While we have to work on these data, I have fit the MM kinetics (1/ substrate vs 1/ rate) based on the initial rate provided by you in the slides that you sent.
- Please find the attached plots. Even here, to fit the MM kinetics I deleted few data points. Here, as you have pointed out when we increase the initial concentration, the rate increases but after a certain limit it decreases. I did not consider the data point after this cut-off limit.
- We need to discuss on the data analysis.
RC (7-29): Do you mean that you and Sudha have used different methods to compute the initial rates and that the results do not agree, or has only Karthik calculated initial rates? I assume you are suggesting some of these experiments need to be repeated?
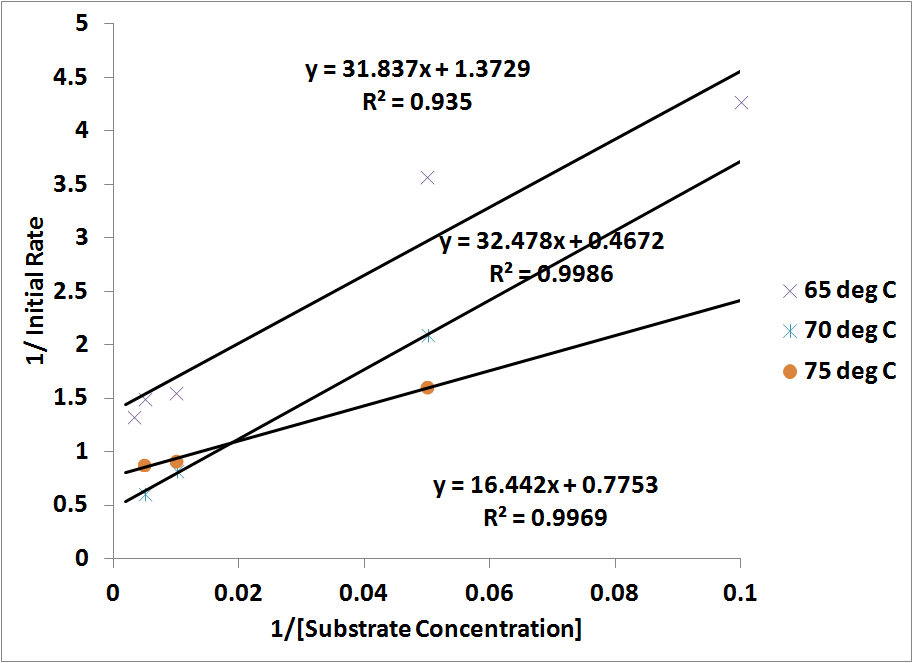
7/13/2013
Minutes of the meeting (Karthik and Sudha).We have discussed the experimental data and below are the conclusion and inference that we made.
1) When the initial concentration of dNTPs increases, the amount of double stranded DNA formed is also increasing. However after some point, increase in initial concentration of dNTPs decreases the amount of DNA formed. This is not an usual observation but according to Sudha this behavior has been already reported in the literature. The initial rate is not yet calculated for all the dNTPs concentions. Once we do this, we need to verify whether all the initial concentration obeys MM kinetics.
2) The calibration curve between RFU and DNA (in ng) is available for 3 units of RFU. Right now it looks like, this is sufficient.
3) The formation of double strand DNA due to the extension reaction has not reached the steady state. Therefore, we need to take this in to account to calculate the initial rate, especially if we use the prism software which does an automatic curve fitting based on the time vs dsDNA data.
4) There are 6 data between time t = 0 to 1 mins and some data does not obey the basic reaction kinetics. For example the dsDNA concentration should always increase from t = 0 but at some time interval this is not the case. So we need to carefully go through these data carefully for all the time interval and remove some of them to improve the quality of the curve fitting. Also we can replace the dsDNA concentration at t = 0, with the dsDNA concentration at t = 0 with no Taq.
5) There are two different trials performed for each suggested experiments. We need to go through these data carefully and calculate the mean value between these two trials or eliminate the data from any particular trial.
6) There is no zero correction done.
7) Sudha will upload the prism data and Karthik will analyze all the data using prism.
7/12/13
Karthik,
Am uploading the experimental results. It is a lot of data, please go through it in brief and I will walk you through it tomorrow.
I have included all the raw data, all the protocols and the calculations wherever possible so that the file is self-explanatory. I realize that the data will take some time to digest but let us discuss any questions you might have with the procedure, calculations etc so that you can work on it while I’m away.
Also, the analyses is necessarily preliminary, as I have mentioned in the last two slides, we need to determine a few critical issues, to understand the data better.
Also if you would like to, you can download Prism (a free demo version is available for 30 days at http://www.graphpad.com/scientific-software/prism/). I can then upload all my Prism files and you will be able to play around with the data analyses as well. Let me know if this appears necessary or feasible.
As we decided last week, let us talk tomorrow, Saturday July 13th at 10.00am.
Sudha.
Taq polymerase activity dNTP varn jun13
6/7/13
Karthik,
I did a test run to check if 2-2000uM dNTP would be a workable range. Please see the attached power point file for results.
Would like you to go thru' this (over the weekend) so we can share thoughts on monday. This file was put together in a rush so please feel free to get back to me with questions by email if you need.
Sudha.
060613 test run
6/4/13
Extension kinetics work discussed during telecon 6/4/13 PM to discuss melt kinetics:
Minutes of telecon 6/4/13 PM (including email exchange leading up to it)
KM: I could understand the Fluorescence data and it says that in trial 1, we could achieve 96% (I have calculated this number based on the initial and final concentration) efficiency in 48 seconds. Whereas in trial 2, the same efficiency was obtained in 40 seconds. This may be probably due to the early melting.
SM:
- 1. I would say that the two trials are quite similar.
- 2. I had incorporated an initial step in the program to measure the fluorescent signal of the ds DNA at the start of the expt. The starting RFU was ~10000. However the melt kinetic curve starts at ~4000. At this point I don’t know whether there was some instantaneous melting (to drop signal from 10000 to 4000 or this is simply a limitation of the PCR software that there is a lack of continuity between cycling steps.
KM: Does the absorbance data also signify the same conclusion?
SM:
- 1. In the UV expt the melting appears slower (it takes ~5-7mins for the curve to plateau as opposed to ~40secs in the PCR machine).
- 2. The curves obtained in the UV expt need smoothing/ fitting.
- 3. At this point I do not know why the Evagreen and Uv methods give different rates of melting. It has to be noted tho’ that the DNA conc in the UV expt is lower and the total volume of sample is higher.
KM: Also, the initial concentration of the DNA is out of the range of typical PCR conditions. I would suggest to use 5nM to 0.5 micro Molar concentration and calculate the melting time. Because, this is the range of the DNA that we encounter in a typical PCR. So we may want to consider to do these experiments.
SM: At this point I cannot devote more time for these expts. My priority is to finish the extension kinetics work.
On the extension experiments:
We should also measure the rate in 45, 55 and 65 deg C.
SM: As per Dr Chakrabarti’s advice, I will assay at 3 temps (50, 60 and 70degC). If reqd, we will then fill the gaps and assay at 3 more temp.s
We have agreed on the following experimental parameters:
Template conc: 200nM
Enzyme conc: 0.36nM
Equilibration : yes: 30mins at assay temp
dNTP conc range: 2, 20, 100, 200, 1000, 2000 uM.
Assay temps: 50, 60, 70oC (initially)
From: Karthikeyan Marimuthu [mailto:[email protected]]
Sent: Tuesday, June 04, 2013 9:44 AM
To: Sudha Moorthy
Subject: RE: extension kinetics
Hi Sudha,
I have understood the following on the Melting experiments results.
There are two different data that are based on Fluorescence and absorbance.
I could understand the Fluorescence data and it says that in trial 1, we could achieve 96% (I have calculated this number based on the initial and final concentration) efficiency in 48 seconds. Whereas in trial 2, the same efficiency was obtained in 40 seconds. This may be probably due to the early melting.
Does the absorbance data also signify the same conclusion?
Also, the initial concentration of the DNA is out of the range of typical PCR conditions. I would suggest to use 5nM to 0.5 micro Molar concentration and calculate the melting time. Because, this is the range of the DNA that we encounter in a typical PCR. So we may want to consider to do these experiments.
On the extension experiments:
We should also measure the rate in 45, 55 and 65 deg C.
I am ok with the second set of concentration. But instead of 1, 2 and 20 micro Molar, we may use 1, 10 and 50 micro Molar.
Karthik.
From: Sudha Moorthy [mailto:[email protected]]
Sent: Monday, June 03, 2013 12:10 PM
To: Karthikeyan Marimuthu
Subject: Re: extension kinetics
Importance: High
Karthik,
Hope you had a chance to take a look at the Lambda DNA melting kinetics results I had posted on Friday.
I am waiting for template DNA (which should arrive by Wednesday) to start the extension kinetics expts., To refresh here are the test conditions:
Template conc: 200nM
Enzyme conc: 0.36nM
Equilibration : yes: 30mins at assay temp
Test Assay temps: 50, 60, 70oC
Would like to confirm with you the dNTP concentrations we are going to use:
I had originally suggested (for purposes of starting the discussion) the following: 20, 50, 100, 200, 500, 1000 uM
However, since you advised that 20uM itself might be excess, I am wondering if one of the following ranges might give us more information:
- 1. 2, 20, 100, 200, 1000, 2000 uM.
- 2. 0.2, 1, 2, 20, 200, 2000 uM
It would be good if you can get back to me regarding this by tomorrow afternoon. In fact if you have time, maybe we can talk tomorrow afternoon or Wednesday morning so that we are agreed on the experimental parameters. Please suggest a convenient time.
Regards,
Sudha.
5/23/13
Minutes of the meeting.
The quality of the extension reaction rate data have been analyzed and discussed (please find the attached ppt for the details of the data).
Based on this analysis, it has been found that this week the reproducibility is quite good.
As per the previous data and discussions we have decided to fix the enzyme concentration to be 0.36 nM and template concentration is 200 nM.
We have decided 6 different nucleotide concentration values (20, 50, 100, 200, 500, 1000 micro molar) at which the initial rate of the extension can be measured.
We have also decided to measure the rate parameters at 6 different temperatures in between 45 to 70 deg C. (45 50 55 60 65 70 deg C). This temperature range is chosen because, the typical PCR is conducted in this temperature range.
Based on the above plan, sudha will make an estimate of amount of reactants required and order them as soon as possible.
5/23/13
Karthik,
Have updated further to include the raw data in the power ppt file itself. Here is the link.
Sudha.
File Not Found
5/23/13
Karthik,
Please see the updated file for our discussion on 5/23/13 PM.
Sudha
File Not Found
5/20/13
Hi Sudha,Please find the attached report on the theoretical part of the extension kinetics paper. I have covered all the contents but it needs a further organization and probably a bit of elaboration. I will do this during this week .
Karthik.
extension paper_theoretical part.docx
5/13/13 Discussion of 5/10/13 results
Hi Sudha,
I have gone through the data carefully and have the following comments.
1) Ideally the assay that was performed without adding any enzyme should show the constant RFU profile, especially for the equilibration case (Because we gaveenough time for the equilibration). The variation that has been observed in the RFU values in this assay should be purely due to the measurement error. The standard deviation in this measurement is 0.36 RFU (with a maximum deviation of 1 RFU). This says that, we can expect the measurement error up to 1 RFU and need to check what will be the corresponding concentration value in moles per liter.
| Picogreen Calibration Eq: |
y = 0.1037x + 0.0103 |
|
| y represents RFU |
||
| x represents ng ds DNA |
||
| When y = 1, x = (1-0.0103)/0.1037 = |
9.54 |
|
| i.e. 1 RFU = |
9.54 |
ng ds DNA |
| = |
4.77 |
ng dNTP incorporated (since only one strand is synthesised) |
| = |
14.71 |
pmols dNTP incorporated |
| = |
1.47E-02 |
nmols dNTP incorporated |
| = |
1.47E-11 |
mols dNTP incorporated |
| -eqbrn |
+eqbrn |
||||||||
| Mins |
+taq |
-taq |
RFU+taq - RFU-taq = DRFU |
zero corrected DRFU |
Mins |
+taq |
-taq |
RFU+taq - RFU-taq = DRFU |
zero corrected DRFU |
| 0.0000 |
8.4074 |
7.0331 |
1.3742 |
0.0000 |
0.0000 |
7.5422 |
6.6607 |
0.8815 |
0.0000 |
| 0.0671 |
8.4698 |
6.9466 |
1.5232 |
0.1489 |
0.0671 |
7.6942 |
6.7010 |
0.9931 |
0.1116 |
2. In the above table, at zeroth minute, the sample that was not equilibrated shows higher RFU values. This shows that some of the double stranded molecules did not melt at 60 deg C. After we gave 30 minutes equilibration time, some double stranded molecues were melted. So, I suspect that the higher RFU values of +taq without equilibration might not be correspond to 60 deg C. Based on this, I think it is safe to proceed with enough equilibration time.
Okay.
3. Also, since we are interested in measuring the initial rate of the reaction, do we need to analyze the data that correspond to 10 minutes. Or Will prism software require this data(up to 10 mins) to calculate the initial rate?. If so, I think in between 2 to 5 minutes, we should add one time interval and also, from 5 to 10 minues we may need to add 2 more time interval. Then only the accuracy of the mean curve that was drawn by the prism software will be good. Do we have any experimental difficulties to include more time intervals after 2 mins (incase if we need RFU values till 10th minute). Otherwise, we should provide only 0 to 2 minutes data to prism to calculate the mean curve and hence the initial rate.
In my initial attempts at data analysis with Prism, I tried several equations to see which would give the best fit and based on this we decided to use the eq. for one-phase association kinetics as follows:

As you can see we need to have data points upto the leveling off of activity so that the curve can be fitted accurately. Also, as we are interested in calculating the initial rate, it is important to define the early time as accurately as possible. If we do not provide Prism with the later values, it cannot fit the data according to One Phase assn kinetics and the fitted curve is not created.
I realize that the more data points we have the better curve we will have, however, over my previous attempts at this assay, we settled on these as the best possible set of minimum data points to use. The main issue with increasing the number of data points is the increased time required for completing the assay. (Remember that each time point is measured individually and each has a 30min pre-incubation before it.)
4. I have also observed that from time t = 0.33 to 2 mins, with taq there is no major difference between RFU values with and without equilibration. This shows that the rate should be the same for first two minutes. After 2 minutes, the assay that was not equilibriated gained more RFU values. I don’t know whether it is because of the experimental error or anything to do with the kinetics. As I have written above if we can have more time intervals then we should be able to get accurate curve that should reduce the prediction error in our kinetic parameter estimation.
I also believe that going with the mean curve data might be not advisable, especially when we don’t have more experimental data from time t = 2 to 10 minutes.
As we have already decided we should repeat this experiment one more time and now with more time intervals.
Here are my suggested experimental parameters:
- 1. Assay temp: 60degC
- 2. Equlibration: yes: 30mins
- 3. No Taq ctrls for each time point: yes
- 4. Time points: 0, 0.33, 0.5, 0.66, 0.83, 1, 2, 3.5, 5, 6.5, 8.5, 10
Sudha.
5/10/13 PM
Karthik,Here is latest update for this week. I am planning for a repeat of this expt for next week. Hopefully we will talk on Monday.
Sudha.
File Not Found
5/10/13

05/10/13
Karthik,Please see attached update for our discussion 5/10/13 11 am. I would like to walk you through the raw data during the discussion and possibly update the results after our meeting.
Sudha.
update 051013
05/03/13
Here is an update on the update.update 050313
05/02/13
Karthik,
Please see attached update. Please feel free to get back with questions when we talk (5/3/13).
Sudha.
Update 050213
04/25/13
Karthik,
Please find attached the plots of extension assays performed at 60deg with/ without equilibration. The equilibration time used was 5mins. The activity with and without equilibration seems to be the same though the activity saturates at a value below that obtained in the 70deg assay.
I am now planning to increase the equilibration time to 20mins and perform assay at 60deg to check if differences in activity can be noted.
Sudha.
042513 update
04/22/13
Karthik,
As I have mentioned before, I would rather not increase Template conc further (the background becomes too high, particularly as the primer is in 7 molar excess as well).
I am planning to assay at 60deg with 5min equilibration and without equilibration.
Sudha.
04/21/2013.
Hi Sudha,After going through the results I have also come to the conclusions that you made.
1) I think 70 deg C is bit high (considering that 80 deg C is a melting temperature) and 5 minutes of equilibration time might have melted some duplexes. Because, for both 0.36 nM and 0.02 nM, though the primer to enzyme ratio is more than 100, the relative rate of melting and enzyme binding decides whether melting dominates the enzyme binding. Typically, enzyme binding is slow and melting is faster. So, this might have affected the enzyme binding reaction and that why 5 mins equilibration gave low RFU values.
2)Also, when the enzyme concentration is 0.36nM for both equilibration and non-equilibration experiments reached a steady state but steady states are different. This shows that the initial concentration of duplex (primer and template) at the time of NTP addition is different for both cases. Further, as I told above the relative rate during the experiment is also important.
3) When the enzyme concentration is 0.02nM with equilibration, the initial concentration of duplex after 5 mins should be equal to whatever the initial concentration of the duplex in the experiment at which 0.36 nM enzyme concentration was maintained with equilibration time. While 0.36nM case reached a steady state, 0.02nM case did not reach the steady state. This not only shows the slow extension reaction but also less enzyme binding.
4) In order to avoid the above scenario, why don't we conduct the assay at 60 or 65 deg C where the primer sequence is very stable. Though the activity of the enzyme at 60 deg C is low compared to 72 deg C, we still can get reasonable activity. So, if possible we can consider 60 deg C first.
5) Or, increase the template and primer and enzyme concentration 10 folds. In this way, we can artificially increase the melting temperature and conduct our kinetic study at 70 deg C.
So based on the above analysis, I would suggest to conduct an experiment at 60 deg C or increase the concentration of enzyme and template both and still maintain the ratio of 1:1000.
04/19/13 PM
Karthik,Also please consider this option:
Will 30 mins equilibration time affect the other parts of the experiments?
The only concern I can think of is as follows:
The Tm of the primer is 80deg. Is it possible that the primer-melting might already have started at 70deg? If that is the case, by equilibrating at 70deg for 30mins do we lose of the primed-template? In fact this could be the reason that the 5min equilibration data is not much higher than the no equilibration data.
We could measure activity at a slightly lower temp, say 65deg. If the primer does not start melting at 65deg, we will not lose activity because of that and 70 to 65deg will not compromise on enzyme activity too much. However, if we choose the lower temp, we will also have to do a comparably no equilibration experiment at 65deg.
Sudha.
04/19/13 PM
Karthik,
Will 30 mins equilibration time affect the other parts of the experiments?
The only concern I can think of is as follows:
The Tm of the primer is 80deg. Is it possible that the primer-melting might already have started at 70deg? If that is the case, by equilibrating at 70deg for 30mins do we lose of the primed-template? In fact this could be the reason that the 5min equilibration data is not much higher than the no equilibration data.
Otherwise, the 30min equilibration will only prolong the experiment time.
I would suggest to conduct an experiment with (only one experiment) with 30 minutes equilibration time.
Even one experiment will need all the time points, otherwise we cannot compare with the 5 min equilibrqtion data and we cannot calculate the initial rate.
Can you also please provide me the rate in number of NTP molecules incorporated/sec/enzyme unit?
To convert mols to no. of molecules, simply multiply the mol by 6.023 X 1023 . Further multiply this number by 100 since the stated rate is for 0.01U Taq. See below:
| Picogreen Calibration Eq#: |
y = 0.1037x + 0.0103 |
|||
| y represents RFU |
||||
| x represents ng ds DNA |
||||
| When y = 1, x = (1-0.0103)/0.1037 = |
9.54 |
|||
| i.e. 1 RFU = |
9.54 |
ng ds DNA |
||
| = |
4.77 |
ng dNTP incorporated (since only one strand is synthesised) |
||
| = |
14.71 |
pmols dNTP incorporated ((ng dNTP*1000)/324.5) (324.5 is the avg mol wt of the dNTPs) |
||
| = |
1.47 X 10-2 |
nmols dNTP incorporated |
||
| = |
1.47 X 10-11 |
mols dNTP incorporated |
||
| dNTP incorporated in 0.2min by 0.01U Taq (+ eqbrn) = |
0.24 RFU* |
= 3.53 X 10-12 pmols |
|
| dNTP incorporated in 1min by 0.01U Taq (+ eqbrn) = |
= 1.76 X 10-11 nmols |
||
| dNTP incorporated in 1sec by 0.01U Taq (+ eqbrn) = |
= 2.94 X 10-13 mols |
||
| dNTP incorporated in 1sec by 1U Taq (+ eqbrn) = |
= 2.94 X 10-11 mols |
||
| = 17.7 X 1012 molecules |
#: This is the previously used calibration curve.
*: This value is obtained from the fitted curve
So, pending further advice from you on Monday, I will plan for the following:
A time course of the extension assay with 20nM template, 0.02nM Taq, assay tem of 70deg, equilibration time of 30mins.
Sudha.
Hi Sudha,
I have quickly gone through the presentation.
As you suggested, though 5 mins equilibration increased the reaction rate, it is no significant. In order to be in safer side, why don’t we increase the equilibration time to, let us say, 30 minutes and compare the reaction rate with 5 mins equilibration time.
Will 30 mins equilibration time affect the other parts of the experiments? If not, I would suggest to conduct an experiment with (only one experiment) with 30 minutes equilibration time.
During the weekend, I will analyze the data carefully and get back to you on the following question
What is there is no significant difference in reaction rate with and without equilibration time.
Can you also please provide me the rate in number of NTP molecules incorporated/sec/enzyme unit?
Karthik.
041813
Karthik,
Please see the attached power point file with results of the preliminary taq polymerase extension assay as per the conditions we have discussed.
It would be good to get a response from you asap so that the next step may be planned. (Preparation and execution of one set of assays takes time.)
- 1.Given the enzyme concentration you recommended, do you expect to see a difference in activity in reactions equilibrated before addition of dNTPs vs reactions that are not equilibrated? (In this trial (5mins equilibration) there appears to be no difference)
- 2.How confident are you that 5mins is sufficient for the equilibration? Since there is no difference between equilibrated and non-equilibrated reactions, would it be worthwhile to try other time periods of equilibration?
- 3.When the reaction conditions are defined, we will have to vary the dNTP conc, so that we can plot the Michaelis-Menten plots and calculate the required parameters. Since the conc I have used here is already in excess, I am guessing that getting the correct range might require a couple of tries.
I have estimated the number of experiments (bare minimum) that would reasonably be required and based on this I have estimated the time required for completion (PROVIDED nothing goes wrong anywhere!) (last slide)
We can, of course, modify this depending on your confidence level on the equilibration time, dNTP conc reqd. etc.
As I mentioned before, please go through the attachment, I need to plan the next steps. If you would like to talk with me regarding results, please feel free to suggest a convenient time (morning is preferable).
Regards,Sudha.
Preliminary Taq Pol Activity Assay Results
040713
Following are minor comments on the MM extension kinetics formulation document below:
- The sentence "During the initial stage of the reaction, assuming there is no [E.Dn] in the product, it is possible to measure the summation of E.D¬i." should be removed.
- Will you be adding this note to the paper? If so, please pass me the edits once you've done them (using the latest version of the paper from Sudha).
- In the paper, we may explicitly state somewhere that [E]_0 \approx [E.Di]_0 (this is currently stated verbally)
- For the paper, I will provide some of my earlier wiki comments regarding the general bireactants formulation, in case we want to present some of Sudha's experiments varying [SP].
Robustness analysis:
Given that the standard MM approach applies to the extension reaction, the novelty of our work will be determined by our presentation of the robustness analysis based on this parameter estimation.
Since we will carry out the robustness analysis under excess nucleotide conditions where the MM formulation is valid, and since you suggest equilibrating SP and enzyme just as we do in this protocol, you should comment further on the wiki regarding why the proposed robustness analysis is not trivial, given that the parameter estimation was done under nearly identical conditions:
a) For one thing, we could interpolate between experimentally studied temperatures (using the Arrhenius formulation activation energy and preexponential).
b) Another suggestion I made was to carry out robustness analysis under variable extension temperature.
c) Finally, bear in mind the following important points regarding the validity of the extension model vis-a-vis initial rates:
- if we want to simulate the entire extension step of a PCR, we cannot assume excess SP for later extension times.
- hence the assumption that all of E is in E.Di form breaks down near the end of the extension step
- for an ordered bireactant reaction, this means that although the second substrate (N) remains in excess throughout, the enzyme in our MM model (here E.Di) eventually becomes "inactive" toward the second substrate due to its conversion to E.Dn (depletion of the first substrate)
- there are two results for later extension times. i) first, we must simulate the enzyme binding step using literature-reported rate constants; ii) second (important), assumptions of the kcat/Kn model break down - constant [ES] was used to derive the kcat/Kn rate law, but [ES] (where S=N) is no longer constant. In our simulations we can follow the evolution of [ES] to determine when the assumption breaks down. Note that the Kapral paper also assumed a constant kcat/Kn in their model.
Due to the issues in c), we may choose to carry out two types of robustness studies - the first is for early extension times, possibly with variable temperature, and the latter is for later extension times, possibly with fixed temperature.
You may disagree with c.i) since you said E.Di cannot dissociate and since the forward rate constant for enzyme binding may be very small at PCR extension temperatures.
Raj
040513
Karthik,
Please see attached file for proposed protocol. Please feel free to edit/ discuss as necessary.
Sudha.
Proposed Protocol to KM 040313.docx
040113 PM
Sudha,
Please see my reply in below in red.
040113 AM
Karthik,
Please see my responses below:
It would be better if the substrate is stable at even 70 deg C. (This will ensure that all enzymes bound on the substrate). Otherwise, we will have to calculate the fraction of the substrate that is available in duplex form at a specific temperature of interest. Accordingly, we can increase the substrate concentration. Essentially, we need to fix the enzyme bound substrate concentration in such a way that there is no free enzyme molecules in the reaction mixture.
SM: Does this mean that the primer-template complex should not dissociate at 70deg?
If yes, this has been taken care of in the design of the template. As you may have noted from the paper, the template is 80bases long with a GC content of 47.5%. The primer binds to a 17mer stretch of bases on one end. This portion has a GC content = 88% and consequently a Tm = 80deg.
KM: If the Tm is 80 deg C, then I am ok with this.
Also, if we use the same enzyme concentration that was used in the previous study (which has a substrate to enzyme concentration ratio to be 100), we can increase the substrate concentration 10 times ( to make the ratio to be 1000). In this case, there would be more enzyme bound substrate molecules which can accept NTPs on it. Therefore, there should be an increase in the Fluorescence signal in this study compared to the previous study.
SM: As I mentioned during our telecon, an expt was conducted with 1uM of substrate and 0.36nM enzyme. The problem encountered was that the background (0 min signal) was too high (note that ~25% of the template oligo is ds at the beginning of the expt). As a consequence, the net gain in RFU was not significant. Please let me know if you would like to see these graphs.
So, my feeling is that the expt will work better if we keep the same substrate conc (20nM) but reduce the enzyme conc further from the previous study (ie make it 0.02nM). Given this, should the nucleotide conc still be the same as before (200uM)?
KM: I assume that in the previous study, the substrate concentration is 20 nM and the enzyme concentration is 0.2 nM (Ratio = 100). We can either increase the substrate concentration to 200 nM or reduce the enzyme concentration to 0.02 nM. I shall leave this to you as you know about the fluorescence signal. Yes, the nucleotide concentration can be fixed to be 200 uM.
Since the previous study might be the case of simultaneous binding and extension reactions( since enzyme and NTPs were added together) which would have anyway consumed all the enzymes if the reaction was conducted for more than 2 mins ( Our PCR simulation suggests this). Therefore, the nucleotide concentration for this new study can be equal to that of previous study.
SM: Don’t know how much/ if at all this will affect your considerations but just wanted to clarify the previously conducted protocol: Briefly, enzyme and nucleotides were NOT added simultaneously.
- The template and primer were annealed in the reaction buffer;
- Enzyme was added to this annealed mix.
- The annealed mix was made as a single master mix for all test conditions to be tested in a day (temp/ time point combinations). The total amount of enzyme reqd for all the reactions was added to this master mix and mixed thoroughly.
- Aliquots of the mix were taken for each test condition e.g., 75deg 10min assay; 75deg 5min assay (and so on) into the PCR tubes placed in the PCR machine. (The master mix was retained at RT while the assays were going on. Would this constitute equilibration? Typically, I start with the longest time point and end with the shortest time point and going thru’ all the time points takes at least 1.5hrs.)
- Reaction was started by addition of nucleotides
- Reaction was stopped by addition of EDTA and chilling. Until all test conditions were tested, the completed assays were stored at 4deg.
- Once all the time points for all the temps were completed, reactions were quantitated with PicoGreen.
Given all this background, I suggest the following:
- Template conc as before.
- Enzyme conc 10 times less
- Include a 5 min equilibration (at room temp?) before addition on nucleotides : KM: You can equilibrate at room temperature (based on how to prepare all the slon) but before adding nucleotide, I would like the reaction mixture to be kept at a temperature of interest for 5 minutes (the temperature at which we measure the reaction kinetics)
- Add the same amount of nucleotides
- I would conduct simultaneously, assays with enzyme conc (same as before) and enzyme conc (10 times less) for one particular temp, say 70deg.
KM: I am with this.
SM: Shall write up the experimental protocol and you can take a look at it.
KM: It would be helpful for me to understand the protocol if you can provide me this and I will comment if I have any questions.
Sudha.
04-01-2013
Hi Sudha,
If the ratio of the Substrate to the enzyme is above 100, then the enzyme binding reaction is quite fast. Therefore, 5 minutes of equilibration time should be sufficient.
It would be better if the substrate is stable at even 70 deg C. (This will ensure that all enzymes bound on the substrate). Otherwise, we will have to calculate the fraction of the substrate that is available in duplex form at a specific temperature of interest. Accordingly, we can increase the substrate concentration. Essentially, we need to fix the enzyme bound substrate concentration in such a way that there is no free enzyme molecules in the reaction mixture.
Also, if we use the same enzyme concentration that was used in the previous study (which has a substrate to enzyme concentration ratio to be 100), we can increase the substrate concentration 10 times ( to make the ratio to be 1000). In this case, there would be more enzyme bound substrate molecules which can accept NTPs on it. Therefore, there should be an increase in the Fluorescence signal in this study compared to the previous study.
For the substrate to enzyme concentration ratio of 100, the equilibrium conversion of the enzyme binding vary from 80 to 50 % ( from 30 to 75 deg C). Since the previous study might be the case of simultaneous binding and extension reactions( since enzyme and NTPs were added together) which would have anyway consumed all the enzymes if the reaction was conducted for more than 2 mins ( Our PCR simulation suggests this). Therefore, the nucleotide concentration for this new study can be equal to that of previous study.
For the substrate to enzyme concentration ratio of 1000, the equilibrium conversion of the enzyme binding vary between 100 to 95% (from 30 to 75 deg C).
To summarize, I would propose the following condition for the first set of experiments.
1) Consider one case of the previous set of experiments at which substrate to enzyme concentration ratio is 100 and there was a good Fluorescence signal.
2) Increase the substrate concentration (SP duplex) by 10 times ( so that the substrate to enzyme concentration ratio is 1000).
3) Fix all other concentration values to be the same.
4) Add the Substrate and enzyme at a specific temperature and maintain the temperature for, let us say 5mins.
5) Add the same amount of NTPs that were added in the previous experiments.
6) Measure the Fluorescence signal as it was done in the previous experiments.
Please let me know if you have any questions.
Best,
Karthik.
Boosalis et al (1987) have used the following concentration values in their study
- 1. Nucleotide concentration was varied from 0 – 8 mM.
- 2. Enzyme concentration was fixed to be approximately 0.2 units in 10 μ ml.
- 3. The estimated ratio between Primer Template = 30:1
- 4. Primer annealing was done separately and then Enzyme was added to it. – This is called as Sol – A.
- 5. Solution B is essentially the NTPs.
- 6. Solution A and B were mixed to conduct the reaction.
03/29/13 Telecon for discussion of
KM(3-28) - Revised Protocol.
Protocol:
1) If the initial SP concentration is 1 μM and the enzyme concentration is 1 nM, we get more than 95% equilibrium conversion and that leaves 10-12 M of Enzyme in the reaction mixture. Here we need to convert the Units of enzyme in to Molar concentration. Typically 1 enzyme unit = 1 μmol/ min.
SM: The experiments reported in the paper were done with 0.5-20nM substrate and 0.36nM enzyme.
A substrate range of 20- 1000nM with 0.36nM enzyme was originally tried. But the assay protocol and the measurement protocol were both tweaked since then. Can go back to take a look at the raw fluorescence numbers; however none of the expts previously conducted have accommodated for equilibration time for enzyme with the substrate.
An expt was tried with 0.18nM enzyme but calc.s were not completed. At the highest substrate conc (20nM), this makes substrate: enzyme 100:1 (your ratio is 1000:1). Can go back to take a look at the raw fluorescence numbers; however as mentioned above, these expts also did not accommodate for equilibration time for enzyme with the substrate.
The reason for taking 0.36nM enzyme is historical; we wanted to be able to compare results with literature.
KM: Calculations suggest that substrate: enzyme of 1000:1 will definitely ensure that all enzyme molecules are bound to substrate.
2) For the above fixed SP and enzyme concentration, Equilibrate the SP and enzymes for long time, so that almost all enzyme molecules bind with the SP molecules.
SM:
How long is a long time?
KM:1- 2hrs??? Dont know how long, I have to think about that.Will give an estimate or upper bound for time reqd.
3) Fix the length of the target DNA to be high (please read the below 3rd comment ).
SM: At this point we have an template oligo that is 80bases long, the primer is 17bases long. So we are looking at an ~60bp extension.
KM: This might be okay. Need to confirm with Raj.
4) Now, add a specific amount of nucleotide with E.SP molecules and measure the rate of the reaction with respect to the time.
SM: When template is 1uM and enzyme is 1nM, what is the estimated nucleotide conc? In the previous expts., nucleotide was taken in excess (200uM).
KM: I don’t know how to fix nucleotide conc. Have papers where polymerase kinetics was done with varying nucleotide conc, will go thru’ them and give an estimate.
Also see SM comments to point 2 below.
5) Repeat the above step for different concentration of Nucleotide
6) The initial concentration of N can be chosen in such a way that it is within a particular limit from the typical nucleotide concentration that was chosen in our previous experiments. In other words, initial concentration of nucleotide can be N +/- ε and ε can be chosen based on experimental convenience.
As mentioned for point (4) in the previous set of expts., nucleotide was taken in excess.
7) Also, in order to check whether the chosen SP concentration is enough to use all the enzyme molecule, assume an another SP concentration which is higher than the original SP concentration and do an extension reaction (for one single N concentration) and compare the Fluorescence data. Now we have Fluorescence data for each SP concentration and compare them. If they are equal, then, the chosen SP concentration can attract all the enzyme molecules.
Comments:
1) Based on the above experiments, the initial rate for various initial concentration of the N is measured.
2) Since N typically in excess compared to the SP molecules, it does not allow E.Di molecules to dissociate back to give Enzyme. Even if there is a dissociation of E from E.Di or E.Dn, those enzyme molecules will bind with the SP molecules ( note that we have excess SP) and produce E.SP. Therefore, at any time during the initial stage of the reaction, all the enzyme molecules will be associated with E.SP and E.Di.
SM: Is my understanding clear that there should be excess substrate and excess nucleotide?
KM: Yes:
Keep substrate conc constant (1uM)
Keep nucleotide conc at what has previously been used (200uM) and then take a range around this conc.
SM: This conc is already excess, so the range will have to be well defined to be able to see a difference in reaction rate.
3) Further, since we measure the initial rate of the reaction we can neglect the concentration of E.Dn molecules. This assumption can be well validated if we can consider an extension reaction with sufficiently a long target DNA ( probably more than 100 base pairs length).
4) Even if the equilibrium between SP and E is shifted because of extension reaction, since the concentration of the free enzyme is almost zero,only very less amount of E.SP molecules only can be produced and their concentration can be neglected.
5) With the above assumptions, it is possible to approximate that the summation of the E.Di and E.Di.N is the initial concentration of the enzyme. Using this, it is possible to derive a typical MM kinetic equation.
Summary
1. It is important that the enzyme conc is much lower than substrate conc so that all enzyme molecules are bound to substrate.
2. It is important to allow for equilibration time for enzyme molecules to bind the substrate molecules.
3. It is okay to start with the nucleotide conc previously used but when we decide to vary the nucleotide conc, the range has to be suitable defined so that differences in initial rate can be measured.
4. A time frame of ~ 3.5 months is estimated: standardizations will be reqd. for the altered substrate:enzyme ratio; for the substrate-enzyme binding equilibration time; and the range of nucleotide conc. range.
5. KM adds: To start with, we will try not to deviate much from the previous experiments protocol by just either increasing the substrate concentration 10 times or reduce the enzyme concentration by 10 times. This will bring the substrate to enzyme concentration ratio to approximately 1000. I will send my calculation with equilibration time and a set of single strand, nucleotide and enzyme concentration values.
KM(3-28)
Notes on the Robustness Analysis.
Measure the total concentration of the nucleotides in duplex form at a specified final time.
We are interested to do the robustness analysis for the extension reaction parameter alone. Therefore, we need to ensure that kinetics of the enzyme binding reaction does not affect the extension reaction kinetics. This can be done by keeping more SP and less enzyme and equilibrate the SP and E before starting the extension reaction.
For a given final time and initial conditions, [E0],[SP0], [N0], [E-SP0], solve the state equations (entire system of coupled extension differential equations.) for the extension reaction. Given the confidence intervals on Km and kcat, we can sample from the uncertainty distribution of parameters to estimate the variance in the total concentration of nucleotides in duplex form. In order to find the variance of the this data, we need to conduct fairly a large number of simulations. I expect that for each simulation, it may take 5-10 minutes. (to enhance the speed we can run the simulation in C++ as well). So, even if we have 100 samples of the rate constants, we should be able to get the variance of total nucleotide concentration in a day.
This can be done in either a fixed or variable temperature framework. An interesting feature of the problem is that there is only one uncertain kinetic parameter.
Another approach to obtaining the total concentration of nucleotides in duplex form at a specified time is to use the Markov chain method you have seen in the attached Kapral paper. This should be equivalent to simulation of the ode system, but may be faster since it doesn't require simulation. It represents the ode system as a single pde. Note that they are able to solve the pde for any temperature profile (time varying rate constant kcat/Km). We should mention this approach in the paper, since otherwise it would appear we are not familiar with it, but we should only use it if we have a problem with simulation time in the robustness analysis.
Another approach to robustness analysis in the (time-varying) linear case is a method based on mechanism identification that I am working on with Andy. We show how for time-varying linear systems one can obtain expressions for the uncertainty distribution without sampling. This is not possible with the Kapral formulation. I won't mention anything more about this for now unless we need it. Again, we may cite it.
I believe that by varying temperature in these extension studies, we can get an idea of whether we will be able to accurately predict complete PCR kinetics in experiments. If we cannot accurately predict extension kinetics, the PCR model may not be sufficiently robust to experimental error. This could be presented as an important motivation for this robustness study.
KM(3-28) - Revised Protocol.
Protocol:
1) If the initial SP concentration is 1 μM and the enzyme concentration is 1 nM, we get more than 95% equilibrium conversion and that leaves 10-12 M of Enzyme in the reaction mixture. Here we need to convert the Units of enzyme in to Molar concentration. Typically 1 enzyme unit = 1 μmol/ min.
2) For the above fixed SP and enzyme concentration, Equilibrate the SP and enzymes for long time, so that almost all enzyme molecules bind with the SP molecules.
3) Fix the length of the target DNA to be high ( please read the below 3rd comment ).
4) Now, add a specific amount of nucleotide with E.SP molecules and measure the rate of the reaction with respect to the time.
5) Repeat the above step for different concentration of Nucleotide
6) The initial concentration of N can be chosen in such a way that it is within a particular limit from the typical nucleotide concentration that was chosen in our previous experiments. In other words, initial concentration of nucleotide can be N +/- ε and ε can be chosen based on experimental convenience.
7) Also, in order to check whether the chosen SP concentration is enough to use all the enzyme molecule, assume one other SP concentration which is higher than the original SP concentration and do an extension reaction (for one single N concentration) and compare the Fluorescence data. Now we have Fluorescence data for each SP concentration and compare them. If they are equal, then, the chosen SP concentration can attract all the enzyme molecules.
Comments:
1) Based on the above experiments, the initial rate for various initial concentration of the N is measured. The time series data (step 4 of the above protocol) that will be obtained for each nucleotide concentration will be fit using the Prism software to estimate the initial rate of the reaction.
2) Since N typically in excess compared to the SP molecules, it does not allow E.Di molecules to dissociate back to give Enzyme. Even if there is a dissociation of E from E.Di or E.Dn, those enzyme molecules will bind with the SP molecules ( note that we have excess SP) and produce E.SP. Therefore, at any time during the initial stage of the reaction, all the enzyme molecules will be associated with E.SP and E.Di.
3) Further, since we measure the initial rate of the reaction we can neglect the concentration of E.Dn molecules. This assumption can be well validated if we can consider an extension reaction with sufficiently a long target DNA ( probably more than 100 base pairs length).
4) With the above assumptions, it is possible to approximate that the summation of the E.Di and E.Di.N is the initial concentration of the enzyme. Using this, it is possible to derive a typical MM kinetic equation.
Best,
Karthik.
03/25/2013
Minutes of Meeting
1. Find an explanation for how to determine the initial rate of the Enzymatic Michaelis Menten (MM) type reaction.
RC (3-25): Not sure this is required anymore, given the use of the prism software to fit the data.
2. Justify that why E.Di can be made constant when the initial rate of the extension reaction is determined.
RC (3-25): Not sure this is required anymore, given the goal of justifying that E.Di is constant in 3.
3. Assume that [SP] molecule is substrate, and [N] is in large excess, and derive an expression for the MM parameters. Also, justify that E.Di are constant in this case (and hence the derivative is zero). Essentially, how the intermediate species concentration is set to a constant in an MM type reaction.
4. Assume [N] is in excess and SP molecules is substrate but the intermediate complex molecule is E.Di.N and derive an equation for the MM parameters.
RC (3-25): Not sure I understand the difference between 3 and 4.
RC (3-25): I think we need to add the task of using the MM formulation with SP as substrate and the MM formulation with N as substrate to determine Km,Kn, and kcat. Including the relationship between kcat's in the two formulations. Also, verification that the MM conditions [E]_0/([S_0]+Km) << 1 are satisfied in the protocols for these formulations.